37 changed files with 961 additions and 84 deletions
Unified View
Diff Options
-
BINcourses/01-dataviz-intro.pdf
-
BINcourses/02-perception-couleurs.pdf
-
BINcourses/03-design-tabulaire.pdf
-
BINcourses/04-interactivite.pdf
-
BINcourses/06_graphes.pdf
-
BINcourses/07_time_text.pdf
-
BINcourses/bar_race.gif
-
+1 -0courses/lab01-correction.Rmd
-
+502 -0courses/lab01-correction.html
-
+0 -3courses/lab01-ggplot-intro.Rmd
-
BINcourses/lab01-ggplot-intro.pdf
-
+56 -11courses/lab02-perception-colors.Rmd
-
+28 -13courses/lab02-perception-colors.html
-
BINcourses/lab02-perception-colors.pdf
-
BINcourses/lab02-perception-colors_files/figure-html/unnamed-chunk-10-1.png
-
BINcourses/lab02-perception-colors_files/figure-html/unnamed-chunk-11-1.png
-
BINcourses/lab02-perception-colors_files/figure-html/unnamed-chunk-13-1.png
-
BINcourses/lab02-perception-colors_files/figure-html/unnamed-chunk-14-1.png
-
BINcourses/lab02-perception-colors_files/figure-html/unnamed-chunk-15-1.png
-
BINcourses/lab02-perception-colors_files/figure-html/unnamed-chunk-7-1.png
-
+6 -6courses/lab03-tabulaire.html
-
BINcourses/lab03-tabulaire.pdf
-
+3 -3courses/lab03_webscraping.html
-
BINcourses/lab03_webscraping.pdf
-
+9 -9courses/lab06_graphes.html
-
BINcourses/lab06_graphes.pdf
-
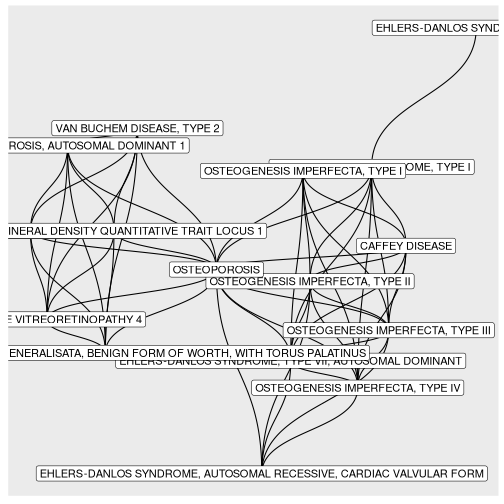
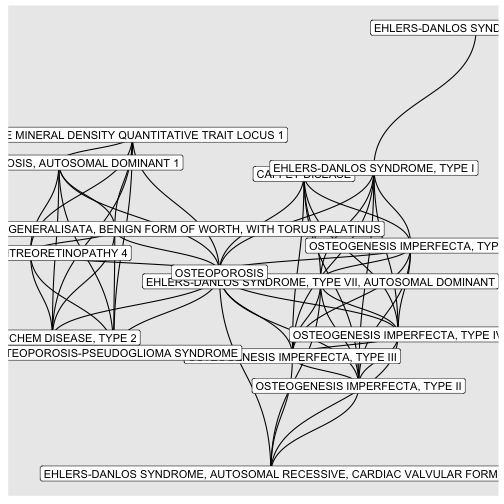
BINcourses/lab06_graphes_files/figure-html/Ostéogénèses-1.png
-
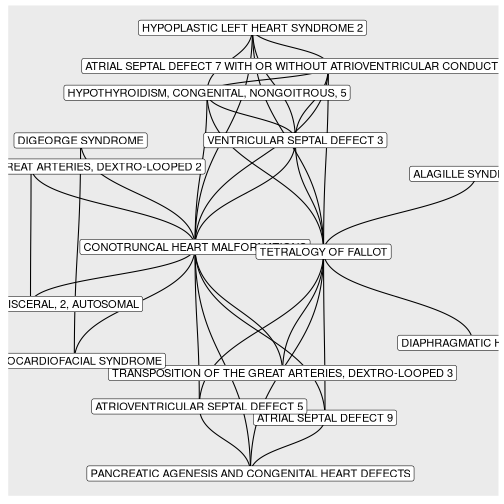
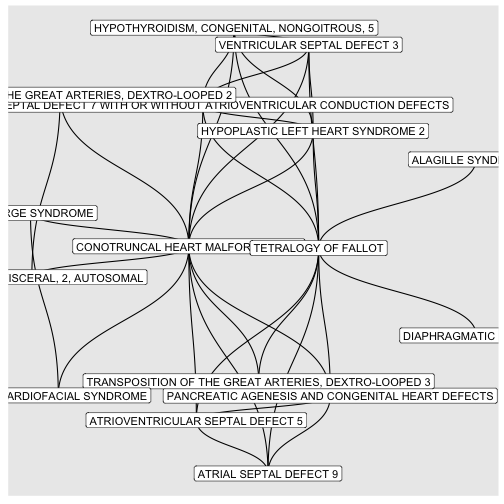
BINcourses/lab06_graphes_files/figure-html/cardiaques-1.png
-
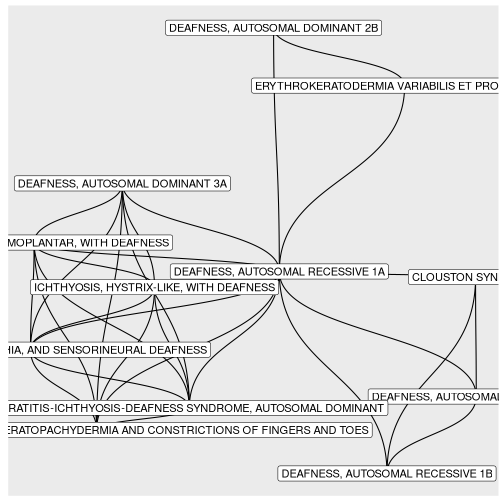
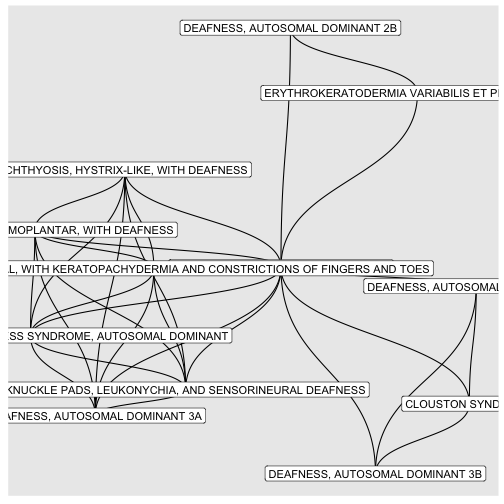
BINcourses/lab06_graphes_files/figure-html/surdités-1.png
-
+137 -20courses/lab7-temporal_data.Rmd
-
+219 -19courses/lab7-temporal_data.html
-
BINcourses/lab7-temporal_data.pdf
-
BINcourses/lab7-temporal_data_files/figure-html/unnamed-chunk-10-1.png
-
BINcourses/lab7-temporal_data_files/figure-html/unnamed-chunk-3-1.png
-
BINcourses/lab7-temporal_data_files/figure-html/unnamed-chunk-4-1.png
-
BINcourses/lab7-temporal_data_files/figure-html/unnamed-chunk-5-1.png
-
BINcourses/lab7-temporal_data_files/figure-html/unnamed-chunk-8-1.png
BIN
courses/01-dataviz-intro.pdf
View File
BIN
courses/02-perception-couleurs.pdf
View File
BIN
courses/03-design-tabulaire.pdf
View File
BIN
courses/04-interactivite.pdf
View File
BIN
courses/06_graphes.pdf
View File
BIN
courses/07_time_text.pdf
View File
BIN
courses/bar_race.gif
View File
+ 1
- 0
courses/lab01-correction.Rmd
View File
| @@ -7,6 +7,7 @@ output: html_document | |||||
| ```{r setup, include=FALSE} | ```{r setup, include=FALSE} | ||||
| knitr::opts_chunk$set(echo = TRUE) | knitr::opts_chunk$set(echo = TRUE) | ||||
| library(ggplot2) | |||||
| ``` | ``` | ||||
| ## Ouvrir le dataset "mtcars" | ## Ouvrir le dataset "mtcars" | ||||
+ 502
- 0
courses/lab01-correction.html
File diff suppressed because it is too large
View File
+ 0
- 3
courses/lab01-ggplot-intro.Rmd
View File
| @@ -13,9 +13,6 @@ output: | |||||
| self-contained: true | self-contained: true | ||||
| beforeInit: "addons/macros.js" | beforeInit: "addons/macros.js" | ||||
| highlightLines: true | highlightLines: true | ||||
| pdf_document: | |||||
| seal: false | |||||
| --- | --- | ||||
| ```{r setup, include=FALSE} | ```{r setup, include=FALSE} | ||||
BIN
courses/lab01-ggplot-intro.pdf
View File
+ 56
- 11
courses/lab02-perception-colors.Rmd
View File
| @@ -25,6 +25,18 @@ library(see) | |||||
| library(RColorBrewer) | library(RColorBrewer) | ||||
| ``` | ``` | ||||
| class: center, middle, title | |||||
| # Lab 2: Perception et couleurs | |||||
| ### 2019-2020 | |||||
| ## Dr. Antoine Neuraz | |||||
| ### AHU Informatique médicale | |||||
| #### Hôpital Necker-Enfants malades, </br> Université de Paris | |||||
| --- | --- | ||||
| class: inverse, center, middle | class: inverse, center, middle | ||||
| # Perception des différentes marques dans ggplot2 | # Perception des différentes marques dans ggplot2 | ||||
| @@ -240,6 +252,14 @@ class: full | |||||
| #### changer la palette par défaut vers une autre palette disponible | #### changer la palette par défaut vers une autre palette disponible | ||||
| --- | |||||
| ```{r} | |||||
| dsamp <- diamonds[sample(nrow(diamonds), 1000), ] | |||||
| ggplot(dsamp, aes(carat, price)) + | |||||
| geom_point(aes(colour = color)) + | |||||
| scale_color_brewer(palette = "Set3") + | |||||
| facet_wrap(~color) | |||||
| ``` | |||||
| --- | --- | ||||
| @@ -254,17 +274,8 @@ class: full | |||||
| #### Caler la palette sur le carat moyen | #### Caler la palette sur le carat moyen | ||||
| #### Annoter le plot avec une ligne désignant le carat moyen et un texte expliquant cette ligne | #### Annoter le plot avec une ligne désignant le carat moyen et un texte expliquant cette ligne | ||||
| --- | --- | ||||
| ```{r} | |||||
| dsamp <- diamonds[sample(nrow(diamonds), 1000), ] | |||||
| ggplot(dsamp, aes(carat, price)) + | |||||
| geom_point(aes(colour = color)) + | |||||
| scale_color_brewer(palette = "Set3") + | |||||
| facet_wrap(~color) | |||||
| ``` | |||||
| ```{r} | ```{r} | ||||
| @@ -273,7 +284,8 @@ ggplot(dsamp, aes(carat, price)) + | |||||
| scale_color_distiller(palette="RdYlBu") | scale_color_distiller(palette="RdYlBu") | ||||
| ``` | ``` | ||||
| ```{r} | |||||
| --- | |||||
| ```{r, eval = F} | |||||
| #showtext_auto() | #showtext_auto() | ||||
| #font_add_google("Schoolbell", "bell") | #font_add_google("Schoolbell", "bell") | ||||
| @@ -307,7 +319,40 @@ ggplot(dsamp, aes(carat, price)) + | |||||
| legend.position = "none") | legend.position = "none") | ||||
| ``` | ``` | ||||
| --- | |||||
| ```{r, echo = F} | |||||
| #showtext_auto() | |||||
| #font_add_google("Schoolbell", "bell") | |||||
| font_family = "sans" | |||||
| annotate_color = "grey50" | |||||
| midpoint = (max(dsamp$carat)-min(dsamp$carat))/2 | |||||
| ggplot(dsamp, aes(carat, price)) + | |||||
| geom_vline(xintercept = midpoint, color = annotate_color) + | |||||
| geom_point(aes(colour = carat)) + | |||||
| scale_color_gradient2(low = "#d8b365", | |||||
| mid="#f5f5f5", | |||||
| high="#5ab4ac", | |||||
| midpoint = midpoint) + | |||||
| annotate("text", | |||||
| x=.78, y=15000, hjust=1, srt=40, | |||||
| label ="this is the midpoint", | |||||
| family=font_family, | |||||
| color=annotate_color) + | |||||
| annotate("curve", | |||||
| x = .8, xend=midpoint-.01, y=15000, yend = 14000, | |||||
| curvature = -.5, | |||||
| color=annotate_color , | |||||
| arrow=arrow(length = unit(0.03, "npc") )) + | |||||
| theme_elegant() + | |||||
| theme(panel.grid.minor = element_blank(), | |||||
| panel.grid.major.x = element_blank(), | |||||
| legend.position = "none") | |||||
| ``` | |||||
+ 28
- 13
courses/lab02-perception-colors.html
View File
| @@ -13,6 +13,18 @@ | |||||
| class: center, middle, title | |||||
| # Lab 2: Perception et couleurs | |||||
| ### 2019-2020 | |||||
| ## Dr. Antoine Neuraz | |||||
| ### AHU Informatique médicale | |||||
| #### Hôpital Necker-Enfants malades, </br> Université de Paris | |||||
| --- | --- | ||||
| class: inverse, center, middle | class: inverse, center, middle | ||||
| # Perception des différentes marques dans ggplot2 | # Perception des différentes marques dans ggplot2 | ||||
| @@ -236,6 +248,19 @@ class: full | |||||
| #### changer la palette par défaut vers une autre palette disponible | #### changer la palette par défaut vers une autre palette disponible | ||||
| --- | |||||
| ```r | |||||
| dsamp <- diamonds[sample(nrow(diamonds), 1000), ] | |||||
| ggplot(dsamp, aes(carat, price)) + | |||||
| geom_point(aes(colour = color)) + | |||||
| scale_color_brewer(palette = "Set3") + | |||||
| facet_wrap(~color) | |||||
| ``` | |||||
| <!-- --> | |||||
| --- | --- | ||||
| ## TODO: couleurs 2 | ## TODO: couleurs 2 | ||||
| @@ -248,20 +273,8 @@ class: full | |||||
| #### Caler la palette sur le carat moyen | #### Caler la palette sur le carat moyen | ||||
| #### Annoter le plot avec une ligne désignant le carat moyen et un texte expliquant cette ligne | #### Annoter le plot avec une ligne désignant le carat moyen et un texte expliquant cette ligne | ||||
| --- | --- | ||||
| ```r | |||||
| dsamp <- diamonds[sample(nrow(diamonds), 1000), ] | |||||
| ggplot(dsamp, aes(carat, price)) + | |||||
| geom_point(aes(colour = color)) + | |||||
| scale_color_brewer(palette = "Set3") + | |||||
| facet_wrap(~color) | |||||
| ``` | |||||
| <!-- --> | |||||
| ```r | ```r | ||||
| @@ -272,6 +285,7 @@ ggplot(dsamp, aes(carat, price)) + | |||||
| <!-- --> | <!-- --> | ||||
| --- | |||||
| ```r | ```r | ||||
| #showtext_auto() | #showtext_auto() | ||||
| @@ -306,7 +320,8 @@ ggplot(dsamp, aes(carat, price)) + | |||||
| legend.position = "none") | legend.position = "none") | ||||
| ``` | ``` | ||||
| <!-- --> | |||||
| --- | |||||
| <!-- --> | |||||
| </textarea> | </textarea> | ||||
| <style data-target="print-only">@media screen {.remark-slide-container{display:block;}.remark-slide-scaler{box-shadow:none;}}</style> | <style data-target="print-only">@media screen {.remark-slide-container{display:block;}.remark-slide-scaler{box-shadow:none;}}</style> | ||||
| <script src="https://remarkjs.com/downloads/remark-latest.min.js"></script> | <script src="https://remarkjs.com/downloads/remark-latest.min.js"></script> | ||||
BIN
courses/lab02-perception-colors.pdf
View File
BIN
courses/lab02-perception-colors_files/figure-html/unnamed-chunk-10-1.png
View File
BIN
courses/lab02-perception-colors_files/figure-html/unnamed-chunk-11-1.png
View File
BIN
courses/lab02-perception-colors_files/figure-html/unnamed-chunk-13-1.png
View File
BIN
courses/lab02-perception-colors_files/figure-html/unnamed-chunk-14-1.png
View File
BIN
courses/lab02-perception-colors_files/figure-html/unnamed-chunk-15-1.png
View File
BIN
courses/lab02-perception-colors_files/figure-html/unnamed-chunk-7-1.png
View File
+ 6
- 6
courses/lab03-tabulaire.html
View File
| @@ -44,8 +44,8 @@ class: center, middle, title | |||||
| read_csv("lab03_data/notes.csv") -> notes | read_csv("lab03_data/notes.csv") -> notes | ||||
| ``` | ``` | ||||
| <div id="htmlwidget-a9f909b507366e2763fd" style="width:100%;height:auto;" class="datatables html-widget"></div> | |||||
| <script type="application/json" data-for="htmlwidget-a9f909b507366e2763fd">{"x":{"filter":"none","data":[["1/1/2019","1/15/2019","2/1/2019","2/15/2019","3/1/2019","3/15/2019","4/1/2019","4/15/2019","5/1/2019","5/15/2019"],[14,16,15,17,14,15,13,15,16,17],[10,11,11,10,15,13,12,12,13,11],[18,19,18,19,19,20,19,19,17,18],[9,10,12,11,14,13,14,15,14,15]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>Date<\/th>\n <th>Alice<\/th>\n <th>Bob<\/th>\n <th>Claire<\/th>\n <th>David<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"paging":false,"info":false,"searching":false,"columnDefs":[{"className":"dt-right","targets":[1,2,3,4]}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> | |||||
| <div id="htmlwidget-94c0f5d1381e56fb5881" style="width:100%;height:auto;" class="datatables html-widget"></div> | |||||
| <script type="application/json" data-for="htmlwidget-94c0f5d1381e56fb5881">{"x":{"filter":"none","data":[["1/1/2019","1/15/2019","2/1/2019","2/15/2019","3/1/2019","3/15/2019","4/1/2019","4/15/2019","5/1/2019","5/15/2019"],[14,16,15,17,14,15,13,15,16,17],[10,11,11,10,15,13,12,12,13,11],[18,19,18,19,19,20,19,19,17,18],[9,10,12,11,14,13,14,15,14,15]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>Date<\/th>\n <th>Alice<\/th>\n <th>Bob<\/th>\n <th>Claire<\/th>\n <th>David<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"paging":false,"info":false,"searching":false,"columnDefs":[{"className":"dt-right","targets":[1,2,3,4]}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> | |||||
| --- | --- | ||||
| @@ -61,8 +61,8 @@ pivot_longer(notes, | |||||
| values_to = "Note") -> notes_long | values_to = "Note") -> notes_long | ||||
| ``` | ``` | ||||
| <div id="htmlwidget-744a3944844d231f7996" style="width:100%;height:auto;" class="datatables html-widget"></div> | |||||
| <script type="application/json" data-for="htmlwidget-744a3944844d231f7996">{"x":{"filter":"none","data":[["1/1/2019","1/1/2019","1/1/2019","1/1/2019","1/15/2019","1/15/2019","1/15/2019","1/15/2019","2/1/2019","2/1/2019","2/1/2019","2/1/2019","2/15/2019","2/15/2019","2/15/2019","2/15/2019","3/1/2019","3/1/2019","3/1/2019","3/1/2019","3/15/2019","3/15/2019","3/15/2019","3/15/2019","4/1/2019","4/1/2019","4/1/2019","4/1/2019","4/15/2019","4/15/2019","4/15/2019","4/15/2019","5/1/2019","5/1/2019","5/1/2019","5/1/2019","5/15/2019","5/15/2019","5/15/2019","5/15/2019"],["Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David"],[14,10,18,9,16,11,19,10,15,11,18,12,17,10,19,11,14,15,19,14,15,13,20,13,13,12,19,14,15,12,19,15,16,13,17,14,17,11,18,15]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>Date<\/th>\n <th>Prénom<\/th>\n <th>Note<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"paging":false,"info":false,"searching":false,"columnDefs":[{"className":"dt-right","targets":2}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> | |||||
| <div id="htmlwidget-3265d78dc11fed97ef02" style="width:100%;height:auto;" class="datatables html-widget"></div> | |||||
| <script type="application/json" data-for="htmlwidget-3265d78dc11fed97ef02">{"x":{"filter":"none","data":[["1/1/2019","1/1/2019","1/1/2019","1/1/2019","1/15/2019","1/15/2019","1/15/2019","1/15/2019","2/1/2019","2/1/2019","2/1/2019","2/1/2019","2/15/2019","2/15/2019","2/15/2019","2/15/2019","3/1/2019","3/1/2019","3/1/2019","3/1/2019","3/15/2019","3/15/2019","3/15/2019","3/15/2019","4/1/2019","4/1/2019","4/1/2019","4/1/2019","4/15/2019","4/15/2019","4/15/2019","4/15/2019","5/1/2019","5/1/2019","5/1/2019","5/1/2019","5/15/2019","5/15/2019","5/15/2019","5/15/2019"],["Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David","Alice","Bob","Claire","David"],[14,10,18,9,16,11,19,10,15,11,18,12,17,10,19,11,14,15,19,14,15,13,20,13,13,12,19,14,15,12,19,15,16,13,17,14,17,11,18,15]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>Date<\/th>\n <th>Prénom<\/th>\n <th>Note<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"paging":false,"info":false,"searching":false,"columnDefs":[{"className":"dt-right","targets":2}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> | |||||
| --- | --- | ||||
| @@ -77,8 +77,8 @@ pivot_wider(notes_long, | |||||
| values_from = Note) | values_from = Note) | ||||
| ``` | ``` | ||||
| <div id="htmlwidget-206da931c15e14bd710f" style="width:100%;height:auto;" class="datatables html-widget"></div> | |||||
| <script type="application/json" data-for="htmlwidget-206da931c15e14bd710f">{"x":{"filter":"none","data":[["1/1/2019","1/15/2019","2/1/2019","2/15/2019","3/1/2019","3/15/2019","4/1/2019","4/15/2019","5/1/2019","5/15/2019"],[14,16,15,17,14,15,13,15,16,17],[10,11,11,10,15,13,12,12,13,11],[18,19,18,19,19,20,19,19,17,18],[9,10,12,11,14,13,14,15,14,15]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>Date<\/th>\n <th>Alice<\/th>\n <th>Bob<\/th>\n <th>Claire<\/th>\n <th>David<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"paging":false,"info":false,"searching":false,"columnDefs":[{"className":"dt-right","targets":[1,2,3,4]}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> | |||||
| <div id="htmlwidget-1496fd73ac98b39c4291" style="width:100%;height:auto;" class="datatables html-widget"></div> | |||||
| <script type="application/json" data-for="htmlwidget-1496fd73ac98b39c4291">{"x":{"filter":"none","data":[["1/1/2019","1/15/2019","2/1/2019","2/15/2019","3/1/2019","3/15/2019","4/1/2019","4/15/2019","5/1/2019","5/15/2019"],[14,16,15,17,14,15,13,15,16,17],[10,11,11,10,15,13,12,12,13,11],[18,19,18,19,19,20,19,19,17,18],[9,10,12,11,14,13,14,15,14,15]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>Date<\/th>\n <th>Alice<\/th>\n <th>Bob<\/th>\n <th>Claire<\/th>\n <th>David<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"paging":false,"info":false,"searching":false,"columnDefs":[{"className":"dt-right","targets":[1,2,3,4]}],"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> | |||||
| --- | --- | ||||
BIN
courses/lab03-tabulaire.pdf
View File
+ 3
- 3
courses/lab03_webscraping.html
File diff suppressed because it is too large
View File
BIN
courses/lab03_webscraping.pdf
View File
+ 9
- 9
courses/lab06_graphes.html
View File
| @@ -233,8 +233,8 @@ graph.star(n = 10, mode = "out") | |||||
| read_csv("lab06_data/OMIM.csv") -> OMIM | read_csv("lab06_data/OMIM.csv") -> OMIM | ||||
| ``` | ``` | ||||
| <div id="htmlwidget-47d834c4dcc97d74e7ee" style="width:100%;height:auto;" class="datatables html-widget"></div> | |||||
| <script type="application/json" data-for="htmlwidget-47d834c4dcc97d74e7ee">{"x":{"filter":"none","data":[["ACYL-CoA DEHYDROGENASE, SHORT-CHAIN, DEFICIENCY OF","ADAMS-OLIVER SYNDROME 1","ADAMS-OLIVER SYNDROME 2","ADAMS-OLIVER SYNDROME 3","ADENINE PHOSPHORIBOSYLTRANSFERASE DEFICIENCY","LUNG CANCER","LUNG CANCER","LUNG CANCER","LUNG CANCER","LUNG CANCER","LUNG CANCER"],["ACADS","ARHGAP31","DOCK6","RBPJ","APRT","CYP2A6","EGFR","TNFSF6","IRF1","BRAF","ERBB2"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>Disease<\/th>\n <th>Gene<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"paging":false,"search":false,"info":false,"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> | |||||
| <div id="htmlwidget-e2536eee24b80cd7b4a4" style="width:100%;height:auto;" class="datatables html-widget"></div> | |||||
| <script type="application/json" data-for="htmlwidget-e2536eee24b80cd7b4a4">{"x":{"filter":"none","data":[["ACYL-CoA DEHYDROGENASE, SHORT-CHAIN, DEFICIENCY OF","ADAMS-OLIVER SYNDROME 1","ADAMS-OLIVER SYNDROME 2","ADAMS-OLIVER SYNDROME 3","ADENINE PHOSPHORIBOSYLTRANSFERASE DEFICIENCY","LUNG CANCER","LUNG CANCER","LUNG CANCER","LUNG CANCER","LUNG CANCER","LUNG CANCER"],["ACADS","ARHGAP31","DOCK6","RBPJ","APRT","CYP2A6","EGFR","TNFSF6","IRF1","BRAF","ERBB2"]],"container":"<table class=\"display\">\n <thead>\n <tr>\n <th>Disease<\/th>\n <th>Gene<\/th>\n <\/tr>\n <\/thead>\n<\/table>","options":{"paging":false,"search":false,"info":false,"order":[],"autoWidth":false,"orderClasses":false}},"evals":[],"jsHooks":[]}</script> | |||||
| --- | --- | ||||
| # Chargement du graphe | # Chargement du graphe | ||||
| @@ -246,9 +246,9 @@ graph.data.frame(OMIM, directed = F) -> graphe | |||||
| ``` | ``` | ||||
| ## IGRAPH 01bad3b UN-- 6288 4234 -- | |||||
| ## IGRAPH d3876e8 UN-- 6288 4234 -- | |||||
| ## + attr: name (v/c) | ## + attr: name (v/c) | ||||
| ## + edges from 01bad3b (vertex names): | |||||
| ## + edges from d3876e8 (vertex names): | |||||
| ## [1] ADRENAL HYPERPLASIA, CONGENITAL, DUE TO 17-ALPHA-HYDROXYLASE DEFICIENCY--CYP17A1 | ## [1] ADRENAL HYPERPLASIA, CONGENITAL, DUE TO 17-ALPHA-HYDROXYLASE DEFICIENCY--CYP17A1 | ||||
| ## [2] 17-BETA-HYDROXYSTEROID DEHYDROGENASE X DEFICIENCY --HSD17B10 | ## [2] 17-BETA-HYDROXYSTEROID DEHYDROGENASE X DEFICIENCY --HSD17B10 | ||||
| ## [3] 2-METHYLBUTYRYL-CoA DEHYDROGENASE DEFICIENCY --ACADSB | ## [3] 2-METHYLBUTYRYL-CoA DEHYDROGENASE DEFICIENCY --ACADSB | ||||
| @@ -304,7 +304,7 @@ neighbors(graphe, V(graphe)[2019]) | |||||
| ``` | ``` | ||||
| ``` | ``` | ||||
| ## + 1/6288 vertex, named, from 01bad3b: | |||||
| ## + 1/6288 vertex, named, from d3876e8: | |||||
| ## [1] SLC25A3 | ## [1] SLC25A3 | ||||
| ``` | ``` | ||||
| @@ -333,9 +333,9 @@ HDN | |||||
| ``` | ``` | ||||
| ``` | ``` | ||||
| ## IGRAPH 8db9c9b UNW- 3512 2839 -- | |||||
| ## IGRAPH 58fb0bd UNW- 3512 2839 -- | |||||
| ## + attr: name (v/c), weight (e/n) | ## + attr: name (v/c), weight (e/n) | ||||
| ## + edges from 8db9c9b (vertex names): | |||||
| ## + edges from 58fb0bd (vertex names): | |||||
| ## [1] 17-BETA-HYDROXYSTEROID DEHYDROGENASE X DEFICIENCY--MENTAL RETARDATION, X-LINKED 17 | ## [1] 17-BETA-HYDROXYSTEROID DEHYDROGENASE X DEFICIENCY--MENTAL RETARDATION, X-LINKED 17 | ||||
| ## [2] 17-BETA-HYDROXYSTEROID DEHYDROGENASE X DEFICIENCY--MENTAL RETARDATION, X-LINKED, SYNDROMIC 10 | ## [2] 17-BETA-HYDROXYSTEROID DEHYDROGENASE X DEFICIENCY--MENTAL RETARDATION, X-LINKED, SYNDROMIC 10 | ||||
| ## [3] 3-HYDROXYACYL-CoA DEHYDROGENASE DEFICIENCY --HYPERINSULINEMIC HYPOGLYCEMIA, FAMILIAL, 4 | ## [3] 3-HYDROXYACYL-CoA DEHYDROGENASE DEFICIENCY --HYPERINSULINEMIC HYPOGLYCEMIA, FAMILIAL, 4 | ||||
| @@ -353,9 +353,9 @@ HGN | |||||
| ``` | ``` | ||||
| ``` | ``` | ||||
| ## IGRAPH 65dfdde UNW- 2776 2810 -- | |||||
| ## IGRAPH f5f35d8 UNW- 2776 2810 -- | |||||
| ## + attr: name (v/c), weight (e/n) | ## + attr: name (v/c), weight (e/n) | ||||
| ## + edges from 65dfdde (vertex names): | |||||
| ## + edges from f5f35d8 (vertex names): | |||||
| ## [1] AKR1C2--AKR1C4 LMNA --MYBPC3 LMNA --ZMPSTE24 GNAS --SSTR5 | ## [1] AKR1C2--AKR1C4 LMNA --MYBPC3 LMNA --ZMPSTE24 GNAS --SSTR5 | ||||
| ## [5] GNAS --AIP GNAS --STX16 GNAS --GNASAS1 COL2A1--COL11A2 | ## [5] GNAS --AIP GNAS --STX16 GNAS --GNASAS1 COL2A1--COL11A2 | ||||
| ## [9] FGFR3 --KRAS FGFR3 --HRAS FGFR3 --RB1 FGFR3 --PIK3CA | ## [9] FGFR3 --KRAS FGFR3 --HRAS FGFR3 --RB1 FGFR3 --PIK3CA | ||||
BIN
courses/lab06_graphes.pdf
View File
BIN
courses/lab06_graphes_files/figure-html/Ostéogénèses-1.png
View File
BIN
courses/lab06_graphes_files/figure-html/cardiaques-1.png
View File
BIN
courses/lab06_graphes_files/figure-html/surdités-1.png
View File
+ 137
- 20
courses/lab7-temporal_data.Rmd
View File
| @@ -21,6 +21,8 @@ library(ggplot2) | |||||
| library(gghighlight) | library(gghighlight) | ||||
| library(dplyr) | library(dplyr) | ||||
| library(ggTimeSeries) | library(ggTimeSeries) | ||||
| library(hrbrthemes) | |||||
| library(gganimate) | |||||
| ``` | ``` | ||||
| ## TODO | ## TODO | ||||
| @@ -47,27 +49,16 @@ library(ggTimeSeries) | |||||
| --- | --- | ||||
| ```{r} | ```{r} | ||||
| data("us_city_populations") | data("us_city_populations") | ||||
| n_cities = 5 | n_cities = 5 | ||||
| # top_cities <- | |||||
| # us_city_populations %>% | |||||
| # filter(Rank <= n_cities) %>% | |||||
| # select(City, State, Region) %>% | |||||
| # distinct() | |||||
| # | |||||
| # to_plot <- filter(us_city_populations, City %in% top_cities$City) | |||||
| #to_plot <- us_city_populations | |||||
| last_ranks <- us_city_populations %>% | last_ranks <- us_city_populations %>% | ||||
| filter(Year == max(Year)) %>% | filter(Year == max(Year)) %>% | ||||
| mutate(last_rank = Rank) %>% | mutate(last_rank = Rank) %>% | ||||
| select(City, last_rank) | select(City, last_rank) | ||||
| to_plot <- left_join(us_city_populations, last_ranks, by= 'City') | |||||
| to_plot <- left_join(us_city_populations, last_ranks, by= 'City') | |||||
| right_axis <- to_plot %>% | right_axis <- to_plot %>% | ||||
| group_by(City) %>% | group_by(City) %>% | ||||
| @@ -85,14 +76,16 @@ labels <- right_axis %>% | |||||
| --- | --- | ||||
| class: full | class: full | ||||
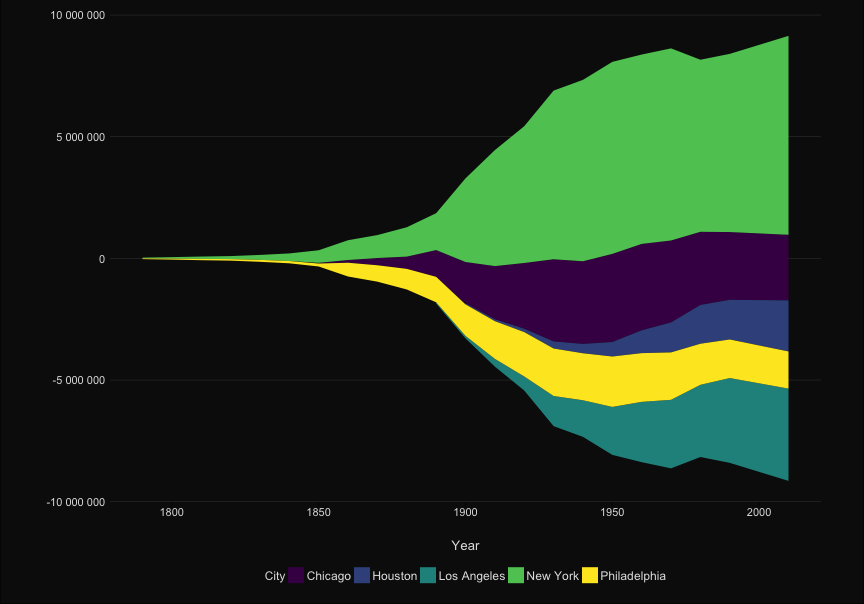
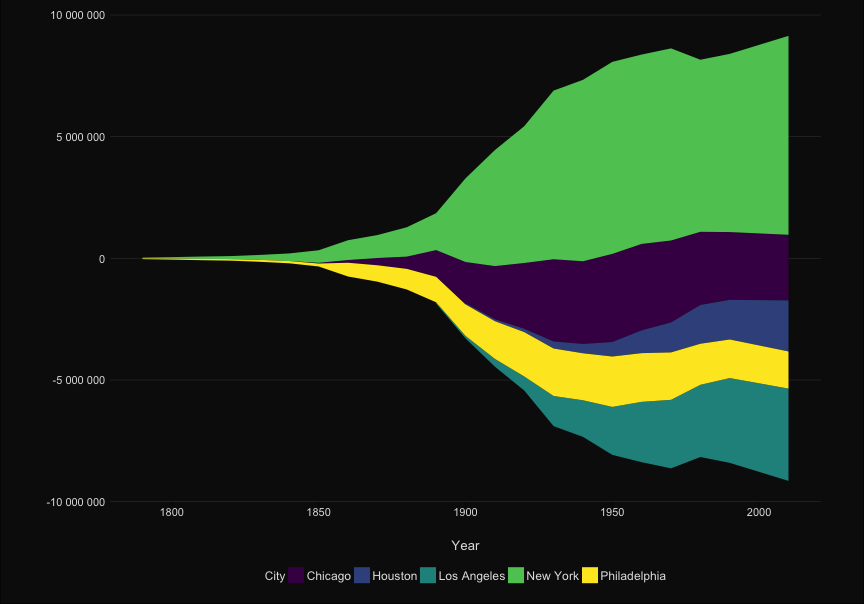
| ```{r, echo = FALSE} | |||||
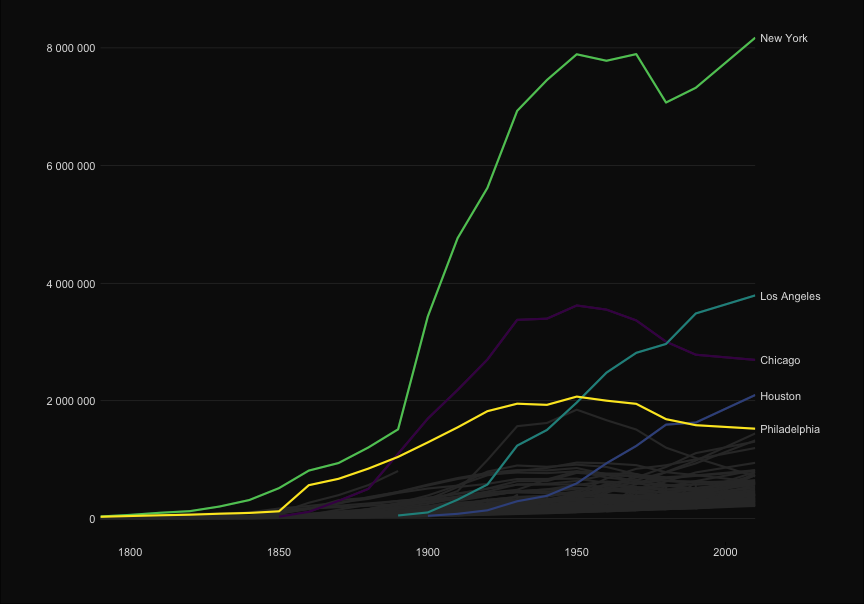
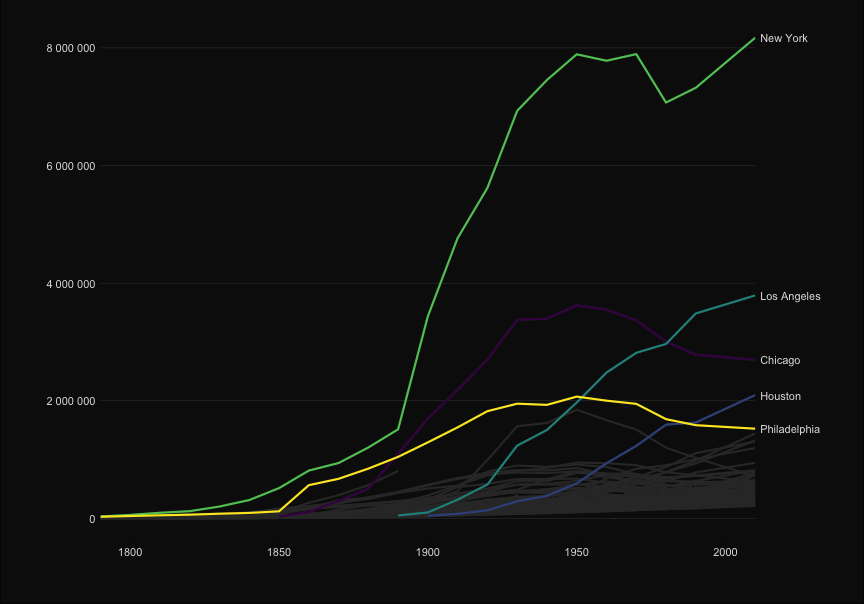
| ggplot(to_plot, aes(x=Year, y = Population, group = City, color = City)) + | |||||
| ```{r, echo = TRUE} | |||||
| p <- ggplot(to_plot, aes(x=Year, y = Population, | |||||
| group = City, color = City)) + | |||||
| geom_line(size=1) + | geom_line(size=1) + | ||||
| #geom_text(data = subset(to_plot, Year == 2010), aes(x=Inf, y = Population, label=City), hjust = 1) + | |||||
| scale_x_continuous("", expand=c(0,0))+ | scale_x_continuous("", expand=c(0,0))+ | ||||
| scale_y_continuous("", | scale_y_continuous("", | ||||
| labels=scales::comma_format(big.mark = " "), | labels=scales::comma_format(big.mark = " "), | ||||
| sec.axis = sec_axis(~ ., breaks = ends, labels = labels ))+ | |||||
| sec.axis = sec_axis(~ ., | |||||
| breaks = ends, | |||||
| labels = labels ))+ | |||||
| scale_color_viridis_d()+ | scale_color_viridis_d()+ | ||||
| theme_elegant_dark()+ | theme_elegant_dark()+ | ||||
| theme(legend.position = "none", | theme(legend.position = "none", | ||||
| @@ -101,14 +94,23 @@ ggplot(to_plot, aes(x=Year, y = Population, group = City, color = City)) + | |||||
| axis.line.x = element_blank(), | axis.line.x = element_blank(), | ||||
| axis.ticks.x = element_line(), | axis.ticks.x = element_line(), | ||||
| panel.grid.major.y = element_line(color= 'grey30', size = .2) ) + | panel.grid.major.y = element_line(color= 'grey30', size = .2) ) + | ||||
| gghighlight(max(last_rank) <= n_cities, use_direct_label = FALSE, label_key = City,unhighlighted_colour = "grey20") | |||||
| gghighlight(max(last_rank) <= n_cities, | |||||
| use_direct_label = FALSE, | |||||
| label_key = City, | |||||
| unhighlighted_colour = "grey20") | |||||
| ``` | ``` | ||||
| --- | --- | ||||
| class: full | class: full | ||||
| ```{r, echo = FALSE} | |||||
| ```{r, echo = TRUE} | |||||
| p | |||||
| ``` | |||||
| --- | |||||
| class: full | |||||
| ```{r, echo = TRUE} | |||||
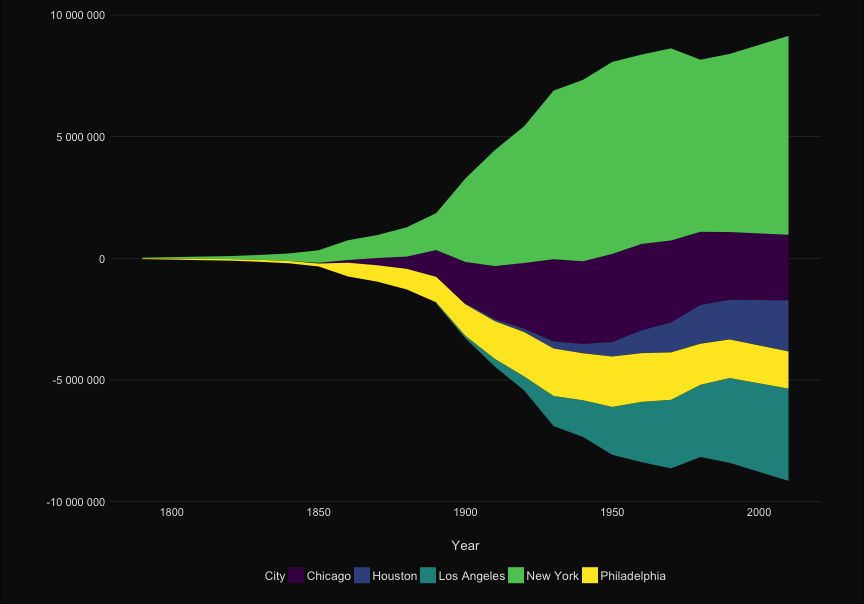
| library(ggTimeSeries) | library(ggTimeSeries) | ||||
| to_plot %>% filter(City %in% labels) %>% | |||||
| p <- to_plot %>% filter(City %in% labels) %>% | |||||
| ggplot(aes(x = Year, y = Population, group = City, fill = City)) + | ggplot(aes(x = Year, y = Population, group = City, fill = City)) + | ||||
| scale_y_continuous("", labels = scales::comma_format(big.mark = " "))+ | scale_y_continuous("", labels = scales::comma_format(big.mark = " "))+ | ||||
| stat_steamgraph() + | stat_steamgraph() + | ||||
| @@ -121,7 +123,122 @@ to_plot %>% filter(City %in% labels) %>% | |||||
| panel.grid.major.y = element_line(color= 'grey30', size = .2) ) | panel.grid.major.y = element_line(color= 'grey30', size = .2) ) | ||||
| ``` | ``` | ||||
| --- | |||||
| class: full | |||||
| ```{r} | |||||
| p | |||||
| ``` | |||||
| --- | --- | ||||
| class: inverse, center, middle | |||||
| # Barchart race | |||||
| --- | |||||
| ## Load data | |||||
| ```{r load_data} | |||||
| data("us_city_populations") | |||||
| n_cities = 10 | |||||
| top_cities <-us_city_populations %>% filter(Rank <= n_cities) %>% | |||||
| select(City, State, Region) %>% distinct() | |||||
| ``` | |||||
| --- | |||||
| ## Create all missing dates | |||||
| ```{r, combine_dates} | |||||
| # create a data frame with all the years between min and max Year | |||||
| all_years <- data.frame(Year = seq(min(us_city_populations$Year), | |||||
| max(us_city_populations$Year), 1)) | |||||
| # combine top_cities and all_years | |||||
| all_combos <- merge(top_cities, all_years, all = T) | |||||
| # combine all_combos with the original dataset | |||||
| res_interp <- merge(us_city_populations, all_combos, all.y = T) | |||||
| ``` | |||||
| ## Interpolate the Populations when missing (linear interpolation here) | |||||
| ```{r, interpolate} | |||||
| res_interp <- res_interp %>% | |||||
| group_by(City) %>% | |||||
| mutate(Population=approx(Year,Population,Year)$y) | |||||
| ``` | |||||
| --- | |||||
| ## Filter data | |||||
| ```{r, filter_for_plot} | |||||
| to_plot <- res_interp %>% | |||||
| group_by(Year) %>% | |||||
| arrange(-Population) %>% | |||||
| mutate(Rank=row_number()) %>% | |||||
| filter(Rank<=n_cities) | |||||
| ``` | |||||
| --- | |||||
| ## Ease transitions | |||||
| ```{r} | |||||
| to_plot_trans <- to_plot %>% | |||||
| group_by(City) %>% | |||||
| arrange(Year) %>% | |||||
| mutate(lag_rank = lag(Rank, 1), | |||||
| change = ifelse(Rank > lag(Rank, 1), 1, 0), | |||||
| change = ifelse(Rank < lag(Rank, 1), -1, 0)) %>% | |||||
| mutate(transition = ifelse(lead(change, 1) == -1, -.9, 0), | |||||
| transition = ifelse(lead(change,2) == -1, -.5, transition), | |||||
| transition = ifelse(lead(change,3) == -1, -.3, transition), | |||||
| transition = ifelse(lead(change, 1) == 1, .9, transition), | |||||
| transition = ifelse(lead(change,2) == 1, .5, transition), | |||||
| transition = ifelse(lead(change,3) == 1, .3, transition)) %>% | |||||
| mutate(trans_rank = Rank + transition) | |||||
| ``` | |||||
| --- | |||||
| ## Make the plot | |||||
| .small[ | |||||
| ```{r, make_plot} | |||||
| p <- to_plot_trans %>% | |||||
| ggplot(aes(x = -trans_rank,y = Population, group =City)) + | |||||
| geom_tile(aes(y = Population / 2, height = Population, fill = Region), | |||||
| width = 0.9) + | |||||
| geom_text(aes(label = City), | |||||
| hjust = "right", colour = "white", | |||||
| fontface="bold", nudge_y = -100000) + | |||||
| geom_text(aes(label = scales::comma(Population,big.mark = ' ')), | |||||
| hjust = "left", nudge_y = 100000, colour = "grey90") + | |||||
| coord_flip(clip="off") + | |||||
| hrbrthemes::scale_fill_ipsum() + | |||||
| scale_x_discrete("") + | |||||
| scale_y_continuous("",labels=scales::comma_format(big.mark = " ")) + | |||||
| theme_elegant_dark(base_size = 20) + | |||||
| theme( | |||||
| panel.grid.minor.x=element_blank(), | |||||
| axis.line = element_blank(), | |||||
| panel.grid.major= element_line(color='lightgrey', size=.2), | |||||
| legend.position = c(0.6, 0.2), | |||||
| plot.margin = margin(1,1,1,2,"cm"), | |||||
| plot.title = element_text(hjust = 0), | |||||
| axis.text.y=element_blank(), | |||||
| legend.text = element_text(size = 15), | |||||
| legend.background = element_blank()) + | |||||
| # gganimate code to transition by year: | |||||
| transition_time(Year) + | |||||
| ease_aes('cubic-in-out') + | |||||
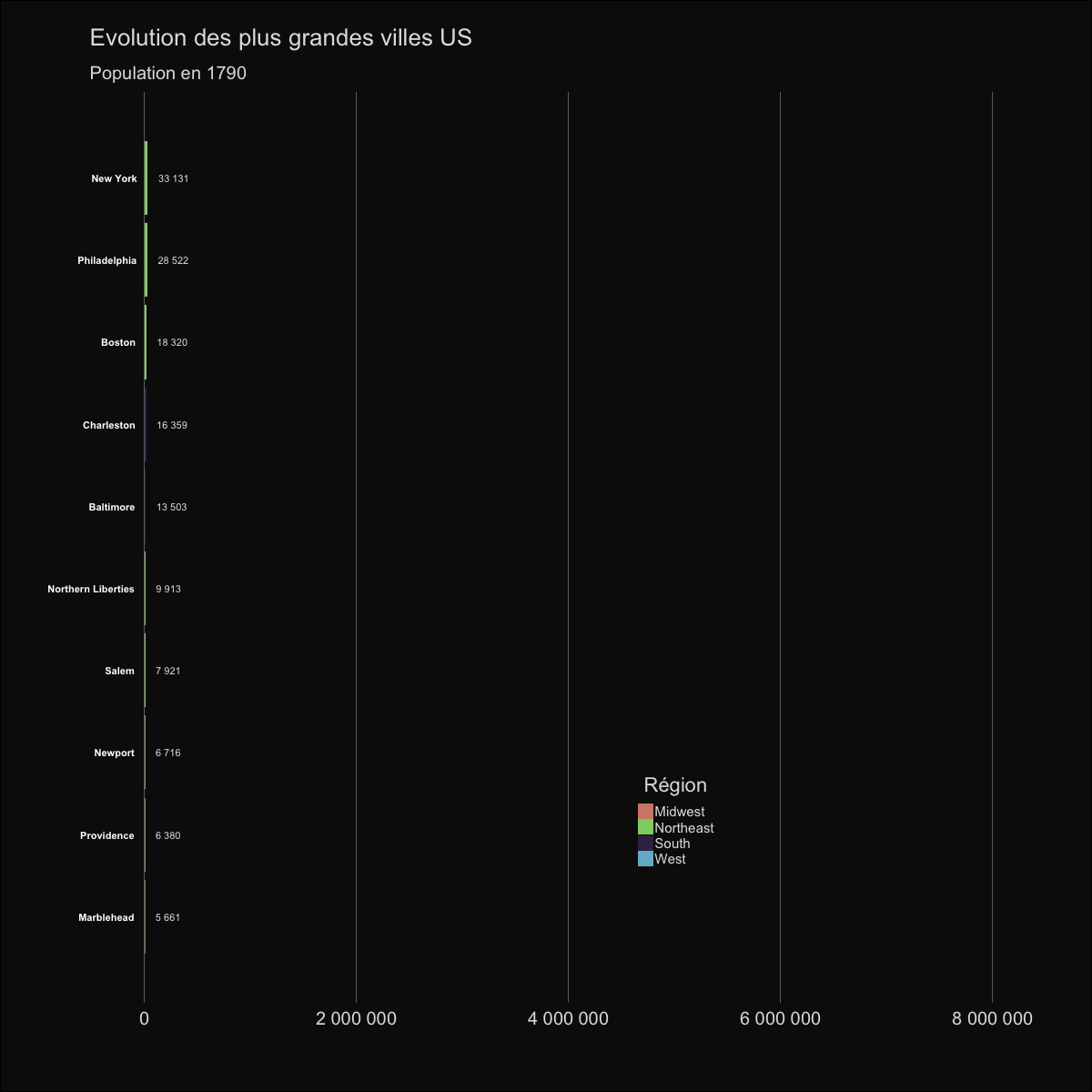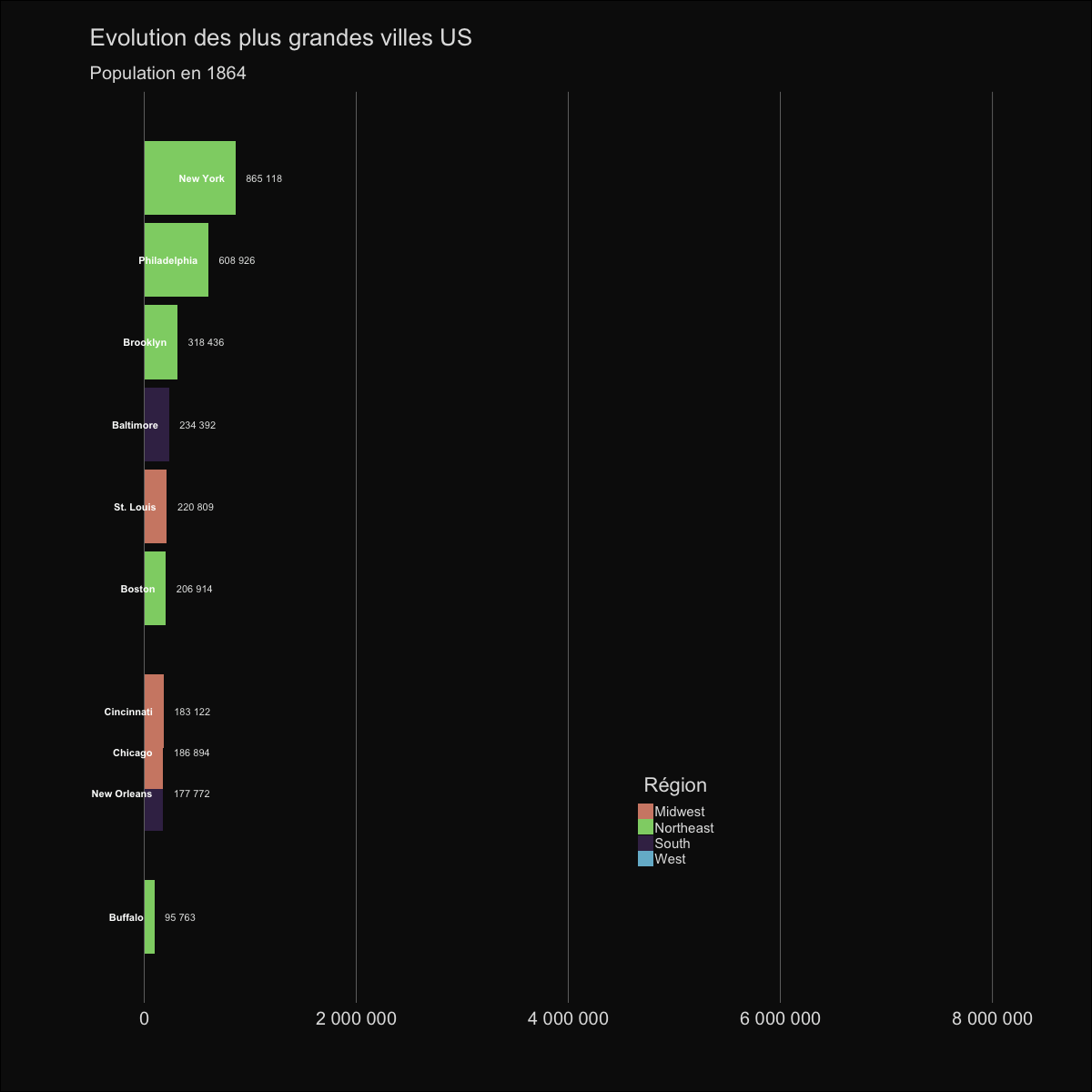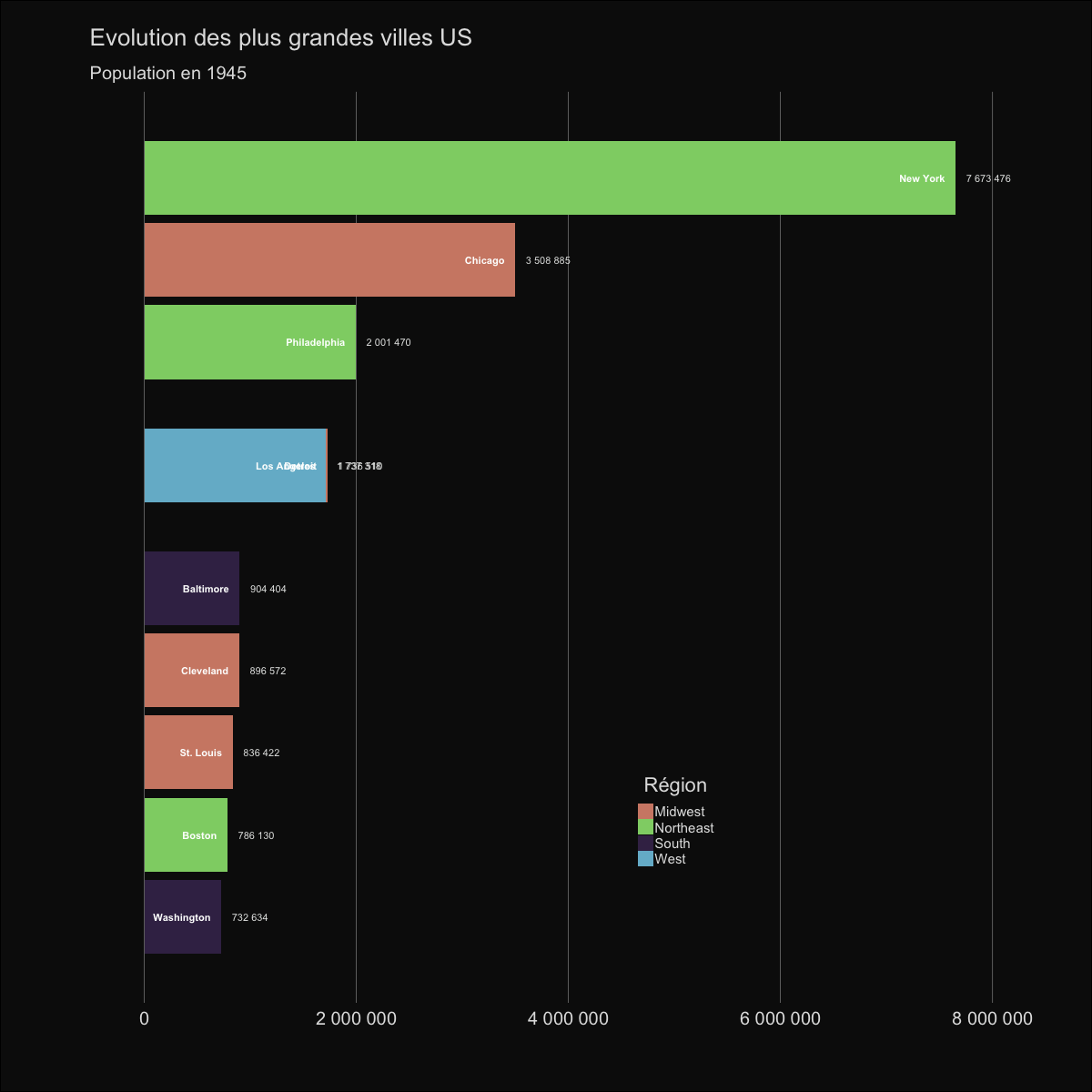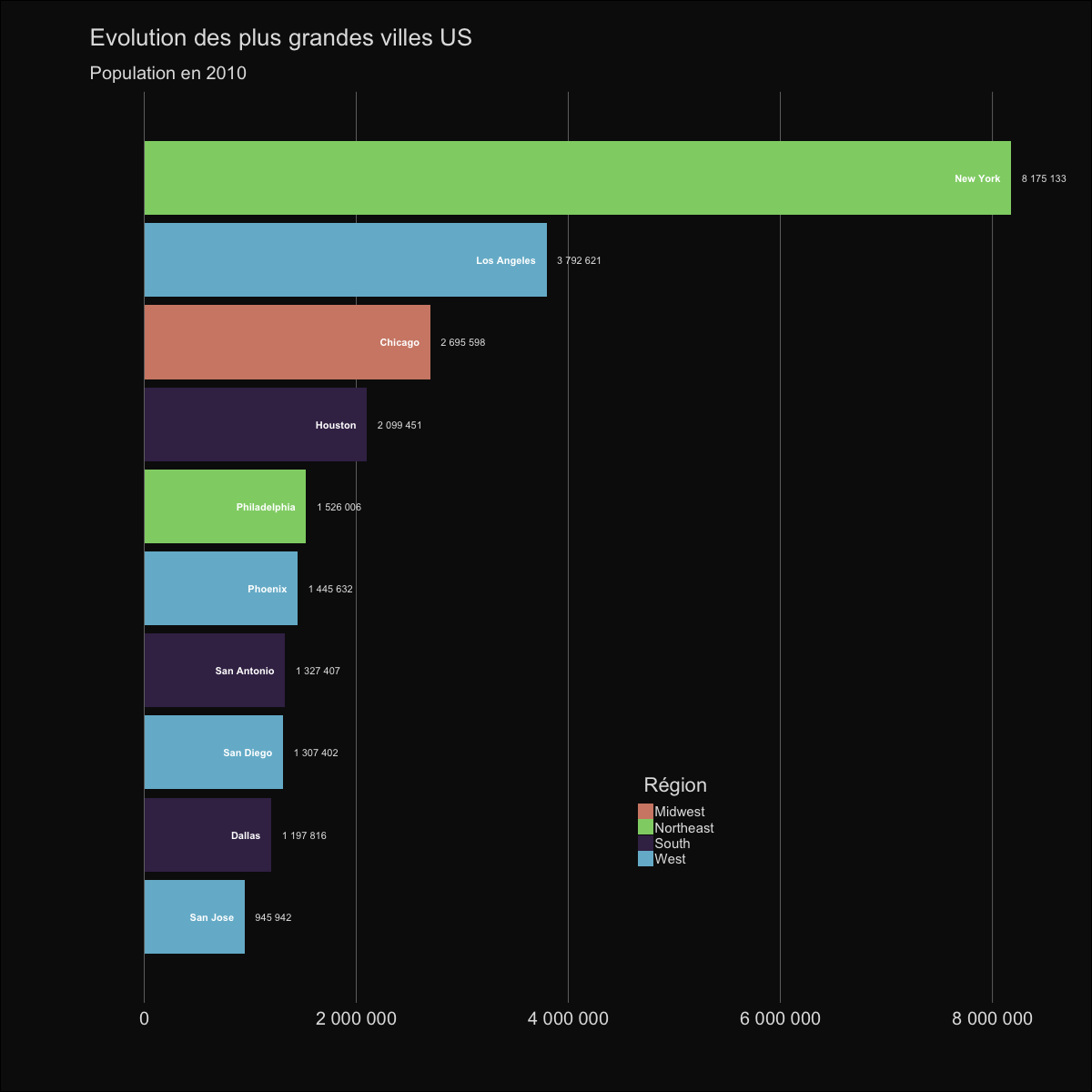
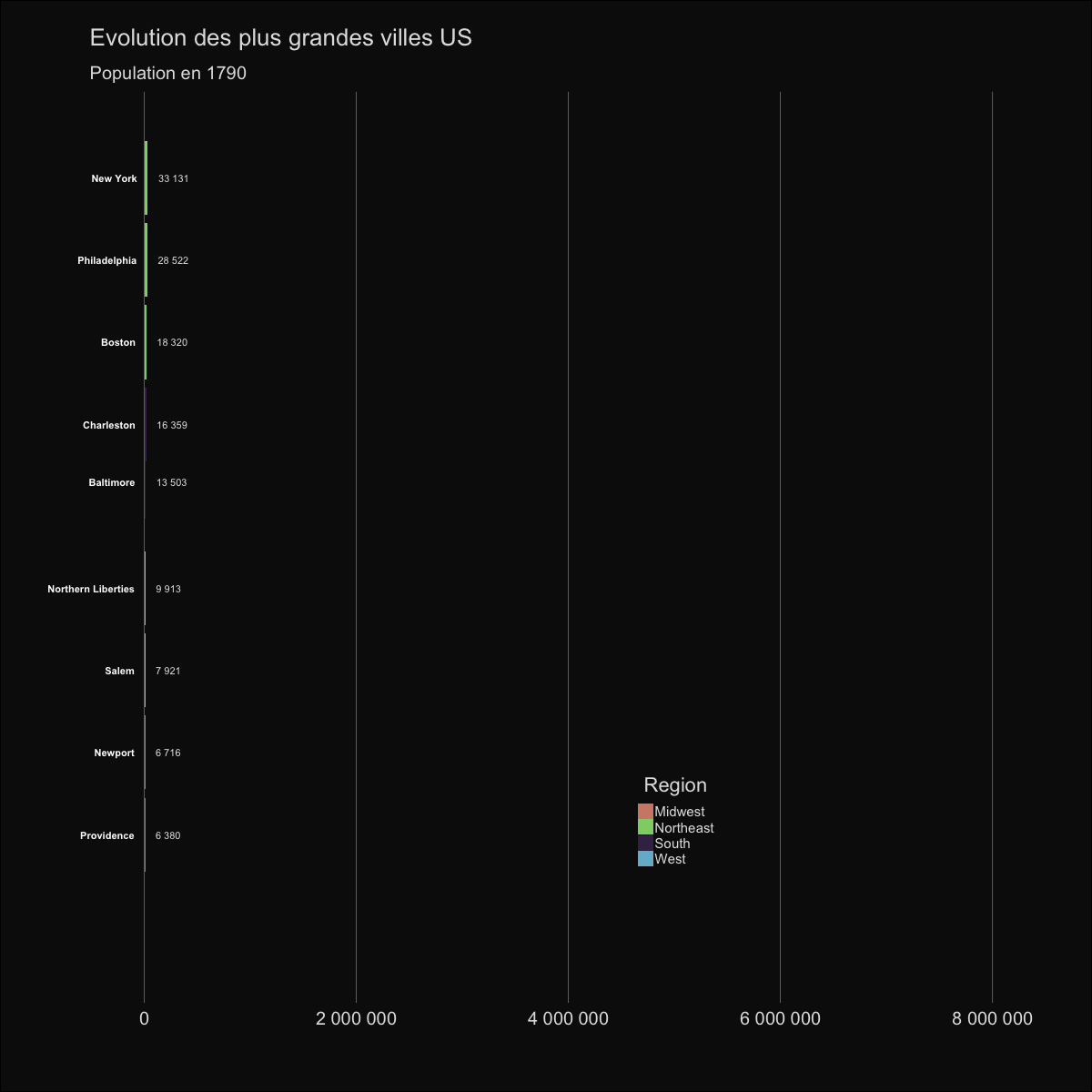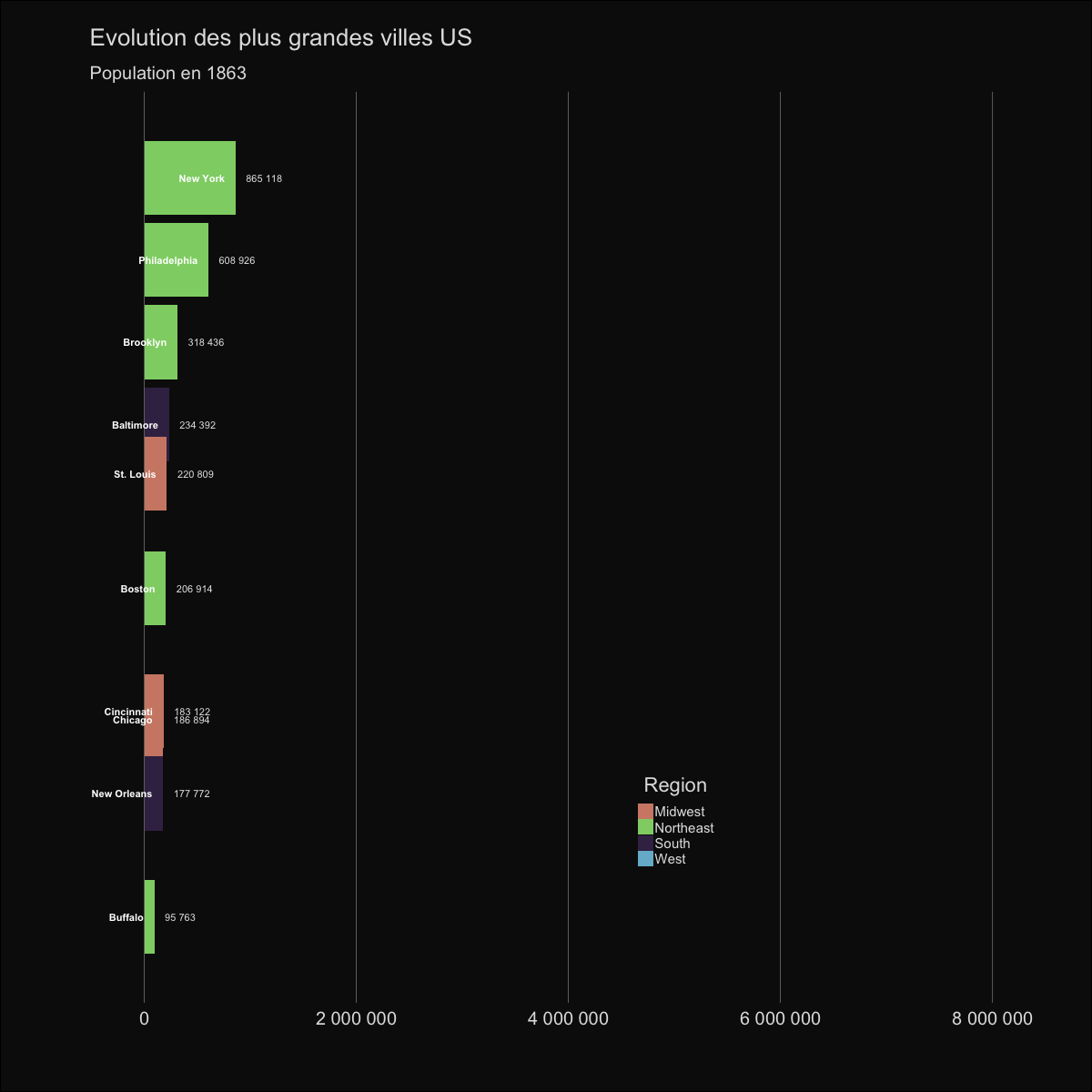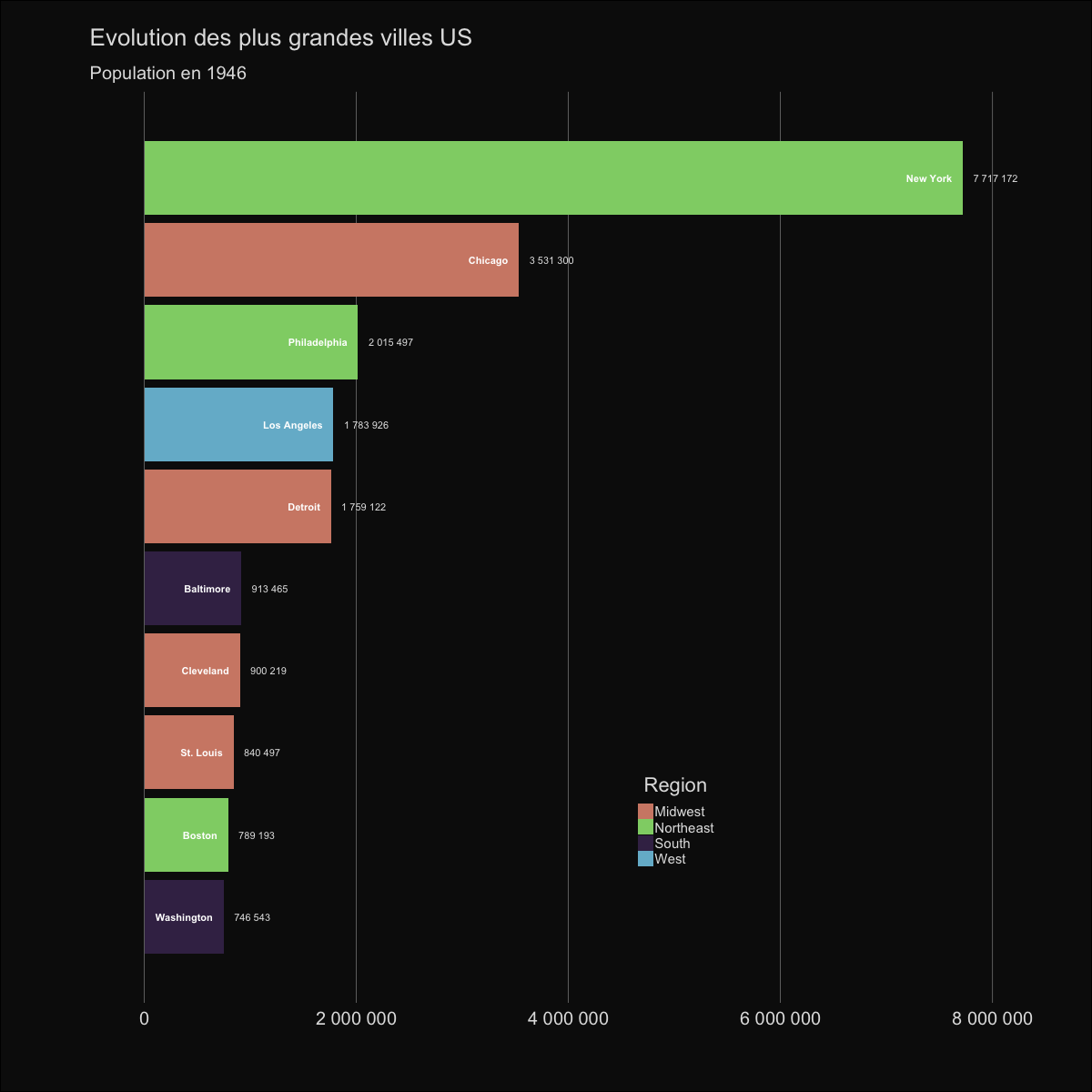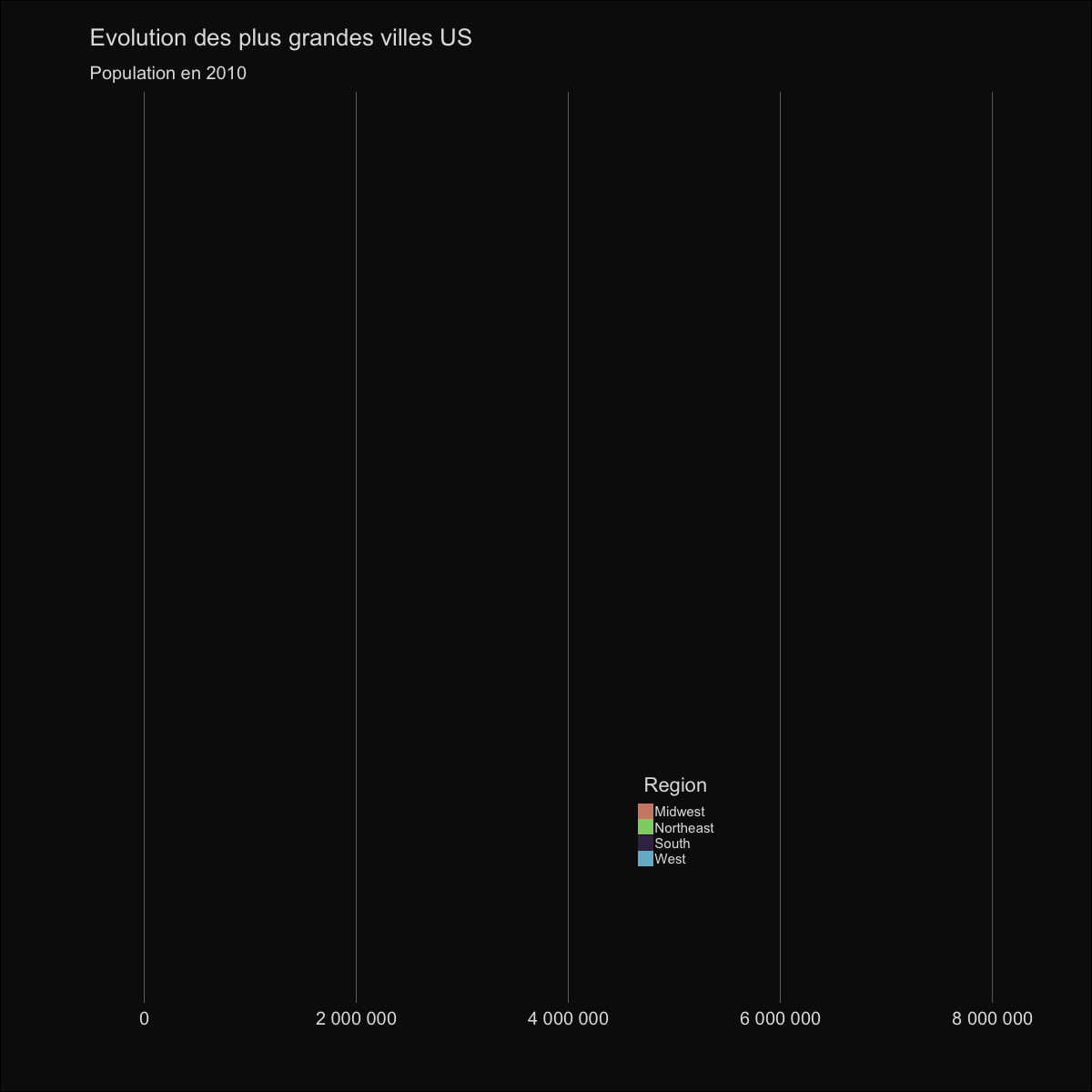
| labs(title='Evolution des plus grandes villes US', | |||||
| subtitle='Population en {round(frame_time,0)}') | |||||
| ``` | |||||
| ] | |||||
| --- | |||||
| ```{r, animate, eval = FALSE} | |||||
| animate(p, nframes = 400, fps = 25, end_pause = 30, width = 1200) | |||||
| anim_save("bar_race.gif", animation = last_animation()) | |||||
| ``` | |||||
|  | |||||
|  | |||||
+ 219
- 19
courses/lab7-temporal_data.html
View File
| @@ -1,8 +1,8 @@ | |||||
| <!DOCTYPE html> | <!DOCTYPE html> | ||||
| <html> | |||||
| <html xmlns="http://www.w3.org/1999/xhtml" lang="" xml:lang=""> | |||||
| <head> | <head> | ||||
| <title>Lab 07 - Données temporelles et textuelles</title> | <title>Lab 07 - Données temporelles et textuelles</title> | ||||
| <meta charset="utf-8"> | |||||
| <meta charset="utf-8" /> | |||||
| <meta name="author" content="Antoine Neuraz" /> | <meta name="author" content="Antoine Neuraz" /> | ||||
| <link href="libs/remark-css-0.0.1/default.css" rel="stylesheet" /> | <link href="libs/remark-css-0.0.1/default.css" rel="stylesheet" /> | ||||
| <link rel="stylesheet" href="css/my_style.css" type="text/css" /> | <link rel="stylesheet" href="css/my_style.css" type="text/css" /> | ||||
| @@ -42,22 +42,12 @@ data("us_city_populations") | |||||
| n_cities = 5 | n_cities = 5 | ||||
| # top_cities <- | |||||
| # us_city_populations %>% | |||||
| # filter(Rank <= n_cities) %>% | |||||
| # select(City, State, Region) %>% | |||||
| # distinct() | |||||
| # | |||||
| # to_plot <- filter(us_city_populations, City %in% top_cities$City) | |||||
| #to_plot <- us_city_populations | |||||
| last_ranks <- us_city_populations %>% | last_ranks <- us_city_populations %>% | ||||
| filter(Year == max(Year)) %>% | filter(Year == max(Year)) %>% | ||||
| mutate(last_rank = Rank) %>% | mutate(last_rank = Rank) %>% | ||||
| select(City, last_rank) | select(City, last_rank) | ||||
| to_plot <- left_join(us_city_populations, last_ranks, by= 'City') | |||||
| to_plot <- left_join(us_city_populations, last_ranks, by= 'City') | |||||
| right_axis <- to_plot %>% | right_axis <- to_plot %>% | ||||
| group_by(City) %>% | group_by(City) %>% | ||||
| @@ -74,14 +64,182 @@ labels <- right_axis %>% | |||||
| --- | --- | ||||
| class: full | class: full | ||||
| <!-- --> | |||||
| ```r | |||||
| p <- ggplot(to_plot, aes(x=Year, y = Population, | |||||
| group = City, color = City)) + | |||||
| geom_line(size=1) + | |||||
| scale_x_continuous("", expand=c(0,0))+ | |||||
| scale_y_continuous("", | |||||
| labels=scales::comma_format(big.mark = " "), | |||||
| sec.axis = sec_axis(~ ., | |||||
| breaks = ends, | |||||
| labels = labels ))+ | |||||
| scale_color_viridis_d()+ | |||||
| theme_elegant_dark()+ | |||||
| theme(legend.position = "none", | |||||
| plot.margin = unit(c(1,3,1,1), "lines"), | |||||
| axis.line.y = element_blank(), | |||||
| axis.line.x = element_blank(), | |||||
| axis.ticks.x = element_line(), | |||||
| panel.grid.major.y = element_line(color= 'grey30', size = .2) ) + | |||||
| gghighlight(max(last_rank) <= n_cities, | |||||
| use_direct_label = FALSE, | |||||
| label_key = City, | |||||
| unhighlighted_colour = "grey20") | |||||
| ``` | |||||
| --- | --- | ||||
| class: full | class: full | ||||
| ```r | |||||
| p | |||||
| ``` | |||||
| <!-- --> | <!-- --> | ||||
| --- | --- | ||||
|  | |||||
| class: full | |||||
| ```r | |||||
| library(ggTimeSeries) | |||||
| p <- to_plot %>% filter(City %in% labels) %>% | |||||
| ggplot(aes(x = Year, y = Population, group = City, fill = City)) + | |||||
| scale_y_continuous("", labels = scales::comma_format(big.mark = " "))+ | |||||
| stat_steamgraph() + | |||||
| theme_elegant_dark() + | |||||
| scale_fill_viridis_d() + | |||||
| theme(plot.margin = unit(c(1,3,1,1), "lines"), | |||||
| axis.line.y = element_blank(), | |||||
| axis.line.x = element_blank(), | |||||
| axis.ticks.x = element_line(), | |||||
| panel.grid.major.y = element_line(color= 'grey30', size = .2) ) | |||||
| ``` | |||||
| --- | |||||
| class: full | |||||
| ```r | |||||
| p | |||||
| ``` | |||||
| <!-- --> | |||||
| --- | |||||
| class: inverse, center, middle | |||||
| # Barchart race | |||||
| --- | |||||
| ## Load data | |||||
| ```r | |||||
| data("us_city_populations") | |||||
| n_cities = 10 | |||||
| top_cities <-us_city_populations %>% filter(Rank <= n_cities) %>% | |||||
| select(City, State, Region) %>% distinct() | |||||
| ``` | |||||
| --- | |||||
| ## Create all missing dates | |||||
| ```r | |||||
| # create a data frame with all the years between min and max Year | |||||
| all_years <- data.frame(Year = seq(min(us_city_populations$Year), | |||||
| max(us_city_populations$Year), 1)) | |||||
| # combine top_cities and all_years | |||||
| all_combos <- merge(top_cities, all_years, all = T) | |||||
| # combine all_combos with the original dataset | |||||
| res_interp <- merge(us_city_populations, all_combos, all.y = T) | |||||
| ``` | |||||
| ## Interpolate the Populations when missing (linear interpolation here) | |||||
| ```r | |||||
| res_interp <- res_interp %>% | |||||
| group_by(City) %>% | |||||
| mutate(Population=approx(Year,Population,Year)$y) | |||||
| ``` | |||||
| --- | |||||
| ## Filter data | |||||
| ```r | |||||
| to_plot <- res_interp %>% | |||||
| group_by(Year) %>% | |||||
| arrange(-Population) %>% | |||||
| mutate(Rank=row_number()) %>% | |||||
| filter(Rank<=n_cities) | |||||
| ``` | |||||
| --- | |||||
| ## Ease transitions | |||||
| ```r | |||||
| to_plot_trans <- to_plot %>% | |||||
| group_by(City) %>% | |||||
| arrange(Year) %>% | |||||
| mutate(lag_rank = lag(Rank, 1), | |||||
| change = ifelse(Rank > lag(Rank, 1), 1, 0), | |||||
| change = ifelse(Rank < lag(Rank, 1), -1, 0)) %>% | |||||
| mutate(transition = ifelse(lead(change, 1) == -1, -.9, 0), | |||||
| transition = ifelse(lead(change,2) == -1, -.5, transition), | |||||
| transition = ifelse(lead(change,3) == -1, -.3, transition), | |||||
| transition = ifelse(lead(change, 1) == 1, .9, transition), | |||||
| transition = ifelse(lead(change,2) == 1, .5, transition), | |||||
| transition = ifelse(lead(change,3) == 1, .3, transition)) %>% | |||||
| mutate(trans_rank = Rank + transition) | |||||
| ``` | |||||
| --- | |||||
| ## Make the plot | |||||
| .small[ | |||||
| ```r | |||||
| p <- to_plot_trans %>% | |||||
| ggplot(aes(x = -trans_rank,y = Population, group =City)) + | |||||
| geom_tile(aes(y = Population / 2, height = Population, fill = Region), | |||||
| width = 0.9) + | |||||
| geom_text(aes(label = City), | |||||
| hjust = "right", colour = "white", | |||||
| fontface="bold", nudge_y = -100000) + | |||||
| geom_text(aes(label = scales::comma(Population,big.mark = ' ')), | |||||
| hjust = "left", nudge_y = 100000, colour = "grey90") + | |||||
| coord_flip(clip="off") + | |||||
| hrbrthemes::scale_fill_ipsum() + | |||||
| scale_x_discrete("") + | |||||
| scale_y_continuous("",labels=scales::comma_format(big.mark = " ")) + | |||||
| theme_elegant_dark(base_size = 20) + | |||||
| theme( | |||||
| panel.grid.minor.x=element_blank(), | |||||
| axis.line = element_blank(), | |||||
| panel.grid.major= element_line(color='lightgrey', size=.2), | |||||
| legend.position = c(0.6, 0.2), | |||||
| plot.margin = margin(1,1,1,2,"cm"), | |||||
| plot.title = element_text(hjust = 0), | |||||
| axis.text.y=element_blank(), | |||||
| legend.text = element_text(size = 15), | |||||
| legend.background = element_blank()) + | |||||
| # gganimate code to transition by year: | |||||
| transition_time(Year) + | |||||
| ease_aes('cubic-in-out') + | |||||
| labs(title='Evolution des plus grandes villes US', | |||||
| subtitle='Population en {round(frame_time,0)}') | |||||
| ``` | |||||
| ] | |||||
| --- | |||||
| ```r | |||||
| animate(p, nframes = 400, fps = 25, end_pause = 30, width = 1200) | |||||
| anim_save("bar_race.gif", animation = last_animation()) | |||||
| ``` | |||||
|  | |||||
| </textarea> | </textarea> | ||||
| <style data-target="print-only">@media screen {.remark-slide-container{display:block;}.remark-slide-scaler{box-shadow:none;}}</style> | |||||
| <script src="https://remarkjs.com/downloads/remark-latest.min.js"></script> | <script src="https://remarkjs.com/downloads/remark-latest.min.js"></script> | ||||
| <script src="addons/macros.js"></script> | <script src="addons/macros.js"></script> | ||||
| <script>var slideshow = remark.create({ | <script>var slideshow = remark.create({ | ||||
| @@ -92,16 +250,57 @@ class: full | |||||
| if (window.HTMLWidgets) slideshow.on('afterShowSlide', function (slide) { | if (window.HTMLWidgets) slideshow.on('afterShowSlide', function (slide) { | ||||
| window.dispatchEvent(new Event('resize')); | window.dispatchEvent(new Event('resize')); | ||||
| }); | }); | ||||
| (function() { | |||||
| var d = document, s = d.createElement("style"), r = d.querySelector(".remark-slide-scaler"); | |||||
| (function(d) { | |||||
| var s = d.createElement("style"), r = d.querySelector(".remark-slide-scaler"); | |||||
| if (!r) return; | if (!r) return; | ||||
| s.type = "text/css"; s.innerHTML = "@page {size: " + r.style.width + " " + r.style.height +"; }"; | s.type = "text/css"; s.innerHTML = "@page {size: " + r.style.width + " " + r.style.height +"; }"; | ||||
| d.head.appendChild(s); | d.head.appendChild(s); | ||||
| })(document); | |||||
| (function(d) { | |||||
| var el = d.getElementsByClassName("remark-slides-area"); | |||||
| if (!el) return; | |||||
| var slide, slides = slideshow.getSlides(), els = el[0].children; | |||||
| for (var i = 1; i < slides.length; i++) { | |||||
| slide = slides[i]; | |||||
| if (slide.properties.continued === "true" || slide.properties.count === "false") { | |||||
| els[i - 1].className += ' has-continuation'; | |||||
| } | |||||
| } | |||||
| var s = d.createElement("style"); | |||||
| s.type = "text/css"; s.innerHTML = "@media print { .has-continuation { display: none; } }"; | |||||
| d.head.appendChild(s); | |||||
| })(document); | |||||
| // delete the temporary CSS (for displaying all slides initially) when the user | |||||
| // starts to view slides | |||||
| (function() { | |||||
| var deleted = false; | |||||
| slideshow.on('beforeShowSlide', function(slide) { | |||||
| if (deleted) return; | |||||
| var sheets = document.styleSheets, node; | |||||
| for (var i = 0; i < sheets.length; i++) { | |||||
| node = sheets[i].ownerNode; | |||||
| if (node.dataset["target"] !== "print-only") continue; | |||||
| node.parentNode.removeChild(node); | |||||
| } | |||||
| deleted = true; | |||||
| }); | |||||
| })();</script> | })();</script> | ||||
| <script> | <script> | ||||
| (function() { | (function() { | ||||
| var i, text, code, codes = document.getElementsByTagName('code'); | |||||
| var links = document.getElementsByTagName('a'); | |||||
| for (var i = 0; i < links.length; i++) { | |||||
| if (/^(https?:)?\/\//.test(links[i].getAttribute('href'))) { | |||||
| links[i].target = '_blank'; | |||||
| } | |||||
| } | |||||
| })(); | |||||
| </script> | |||||
| <script> | |||||
| slideshow._releaseMath = function(el) { | |||||
| var i, text, code, codes = el.getElementsByTagName('code'); | |||||
| for (i = 0; i < codes.length;) { | for (i = 0; i < codes.length;) { | ||||
| code = codes[i]; | code = codes[i]; | ||||
| if (code.parentNode.tagName !== 'PRE' && code.childElementCount === 0) { | if (code.parentNode.tagName !== 'PRE' && code.childElementCount === 0) { | ||||
| @@ -115,7 +314,8 @@ if (window.HTMLWidgets) slideshow.on('afterShowSlide', function (slide) { | |||||
| } | } | ||||
| i++; | i++; | ||||
| } | } | ||||
| })(); | |||||
| }; | |||||
| slideshow._releaseMath(document); | |||||
| </script> | </script> | ||||
| <!-- dynamically load mathjax for compatibility with self-contained --> | <!-- dynamically load mathjax for compatibility with self-contained --> | ||||
| <script> | <script> | ||||