共有 15 个文件被更改,包括 6325 次插入 和 0 次删除
分列视图
Diff 选项
-
二进制courses/06_img/71.png
-
二进制courses/06_img/72.png
-
二进制courses/06_img/73.png
-
二进制courses/06_img/74.png
-
二进制courses/06_img/75.png
-
二进制courses/06_img/76.png
-
二进制courses/06_img/77.png
-
二进制courses/06_img/HDN.png
-
二进制courses/06_img/HDN_principe.png
-
二进制courses/06_img/bipartite.png
-
+1525 -0courses/06_img/bipartite.svg
-
二进制courses/06_img/projection.png
-
+191 -0courses/06_img/projection.svg
-
+4235 -0courses/lab06_data/OMIM.csv
-
+374 -0courses/lab06_graphes.Rmd
二进制
courses/06_img/71.png
查看文件
二进制
courses/06_img/72.png
查看文件
二进制
courses/06_img/73.png
查看文件
二进制
courses/06_img/74.png
查看文件
二进制
courses/06_img/75.png
查看文件
二进制
courses/06_img/76.png
查看文件
二进制
courses/06_img/77.png
查看文件
二进制
courses/06_img/HDN.png
查看文件
二进制
courses/06_img/HDN_principe.png
查看文件
二进制
courses/06_img/bipartite.png
查看文件
+ 1525
- 0
courses/06_img/bipartite.svg
文件差异内容过多而无法显示
查看文件
二进制
courses/06_img/projection.png
查看文件
+ 191
- 0
courses/06_img/projection.svg
查看文件
| @@ -0,0 +1,191 @@ | |||
| <?xml version="1.0" encoding="UTF-8" standalone="no"?> | |||
| <!-- Created with Inkscape (http://www.inkscape.org/) --> | |||
| <svg | |||
| xmlns:dc="http://purl.org/dc/elements/1.1/" | |||
| xmlns:cc="http://creativecommons.org/ns#" | |||
| xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#" | |||
| xmlns:svg="http://www.w3.org/2000/svg" | |||
| xmlns="http://www.w3.org/2000/svg" | |||
| xmlns:sodipodi="http://sodipodi.sourceforge.net/DTD/sodipodi-0.dtd" | |||
| xmlns:inkscape="http://www.inkscape.org/namespaces/inkscape" | |||
| width="210mm" | |||
| height="297mm" | |||
| viewBox="0 0 210 297" | |||
| version="1.1" | |||
| id="svg8" | |||
| inkscape:version="0.92.4 5da689c313, 2019-01-14" | |||
| sodipodi:docname="projection.svg" | |||
| inkscape:export-filename="/home/maxx/Documents/Cours/dataviz/courses/06_img/projection.png" | |||
| inkscape:export-xdpi="100" | |||
| inkscape:export-ydpi="100"> | |||
| <defs | |||
| id="defs2" /> | |||
| <sodipodi:namedview | |||
| id="base" | |||
| pagecolor="#ffffff" | |||
| bordercolor="#666666" | |||
| borderopacity="1.0" | |||
| inkscape:pageopacity="0.0" | |||
| inkscape:pageshadow="2" | |||
| inkscape:zoom="1.4" | |||
| inkscape:cx="-90.551175" | |||
| inkscape:cy="351.95387" | |||
| inkscape:document-units="mm" | |||
| inkscape:current-layer="layer1" | |||
| showgrid="false" /> | |||
| <metadata | |||
| id="metadata5"> | |||
| <rdf:RDF> | |||
| <cc:Work | |||
| rdf:about=""> | |||
| <dc:format>image/svg+xml</dc:format> | |||
| <dc:type | |||
| rdf:resource="http://purl.org/dc/dcmitype/StillImage" /> | |||
| <dc:title></dc:title> | |||
| </cc:Work> | |||
| </rdf:RDF> | |||
| </metadata> | |||
| <g | |||
| inkscape:label="Layer 1" | |||
| inkscape:groupmode="layer" | |||
| id="layer1"> | |||
| <g | |||
| id="g8382" | |||
| transform="translate(-161.86943,55.327505)"> | |||
| <circle | |||
| style="display:inline;opacity:1;fill:#ffffff;fill-opacity:1;stroke:#000000;stroke-width:0.35277778;stroke-miterlimit:10;stroke-dasharray:none;stroke-opacity:1" | |||
| id="circle827-8" | |||
| cx="135.72226" | |||
| cy="105.37312" | |||
| r="4.0497961" /> | |||
| <text | |||
| id="text7175-7" | |||
| y="107.40246" | |||
| x="133.81709" | |||
| style="font-style:normal;font-variant:normal;font-weight:normal;font-stretch:normal;font-size:7.76111126px;line-height:1.25;font-family:'Trebuchet MS';-inkscape-font-specification:'Trebuchet MS, Normal';font-variant-ligatures:normal;font-variant-caps:normal;font-variant-numeric:normal;font-feature-settings:normal;text-align:start;letter-spacing:0px;word-spacing:0px;writing-mode:lr-tb;text-anchor:start;display:inline;fill:#000000;fill-opacity:1;stroke:none;stroke-width:0.26458332" | |||
| xml:space="preserve"><tspan | |||
| style="font-size:7.76111126px;stroke-width:0.26458332" | |||
| y="107.40246" | |||
| x="133.81709" | |||
| id="tspan7173-92" | |||
| sodipodi:role="line">a</tspan></text> | |||
| </g> | |||
| <g | |||
| id="g8405" | |||
| transform="translate(-129.22985,-17.914504)"> | |||
| <circle | |||
| r="4.0497961" | |||
| cy="179.67406" | |||
| cx="135.72226" | |||
| id="circle825-2" | |||
| style="display:inline;opacity:1;fill:#ffffff;fill-opacity:1;stroke:#000000;stroke-width:0.35277778;stroke-miterlimit:10;stroke-dasharray:none;stroke-opacity:1" /> | |||
| <text | |||
| id="text7175-3-3" | |||
| y="182.36557" | |||
| x="133.41588" | |||
| style="font-style:normal;font-variant:normal;font-weight:normal;font-stretch:normal;font-size:7.76111126px;line-height:1.25;font-family:'Trebuchet MS';-inkscape-font-specification:'Trebuchet MS, Normal';font-variant-ligatures:normal;font-variant-caps:normal;font-variant-numeric:normal;font-feature-settings:normal;text-align:start;letter-spacing:0px;word-spacing:0px;writing-mode:lr-tb;text-anchor:start;display:inline;fill:#000000;fill-opacity:1;stroke:none;stroke-width:0.26458332" | |||
| xml:space="preserve"><tspan | |||
| style="font-size:7.76111126px;stroke-width:0.26458332" | |||
| y="182.36557" | |||
| x="133.41588" | |||
| id="tspan7173-5-7" | |||
| sodipodi:role="line">d</tspan></text> | |||
| </g> | |||
| <g | |||
| id="g8387" | |||
| transform="translate(-134.84413,53.278064)"> | |||
| <circle | |||
| style="display:inline;opacity:1;fill:#ffffff;fill-opacity:1;stroke:#000000;stroke-width:0.35277778;stroke-miterlimit:10;stroke-dasharray:none;stroke-opacity:1" | |||
| id="circle2450-7" | |||
| cx="135.72226" | |||
| cy="130.14009" | |||
| r="4.0497961" /> | |||
| <text | |||
| id="text7175-9-3" | |||
| y="132.96335" | |||
| x="133.71387" | |||
| style="font-style:normal;font-variant:normal;font-weight:normal;font-stretch:normal;font-size:7.76111126px;line-height:1.25;font-family:'Trebuchet MS';-inkscape-font-specification:'Trebuchet MS, Normal';font-variant-ligatures:normal;font-variant-caps:normal;font-variant-numeric:normal;font-feature-settings:normal;text-align:start;letter-spacing:0px;word-spacing:0px;writing-mode:lr-tb;text-anchor:start;display:inline;fill:#000000;fill-opacity:1;stroke:none;stroke-width:0.26458332" | |||
| xml:space="preserve"><tspan | |||
| style="font-size:7.76111126px;stroke-width:0.26458332" | |||
| y="132.96335" | |||
| x="133.71387" | |||
| id="tspan7173-1-6" | |||
| sodipodi:role="line">b</tspan></text> | |||
| </g> | |||
| <g | |||
| id="g8392" | |||
| transform="translate(-153.17598,34.105708)"> | |||
| <circle | |||
| r="4.0497961" | |||
| cy="154.90707" | |||
| cx="135.72226" | |||
| id="circle823-9" | |||
| style="display:inline;opacity:1;fill:#ffffff;fill-opacity:1;stroke:#000000;stroke-width:0.35277778;stroke-miterlimit:10;stroke-dasharray:none;stroke-opacity:1" /> | |||
| <text | |||
| id="text7175-0-4" | |||
| y="156.9364" | |||
| x="133.6673" | |||
| style="font-style:normal;font-variant:normal;font-weight:normal;font-stretch:normal;font-size:7.76111126px;line-height:1.25;font-family:'Trebuchet MS';-inkscape-font-specification:'Trebuchet MS, Normal';font-variant-ligatures:normal;font-variant-caps:normal;font-variant-numeric:normal;font-feature-settings:normal;text-align:start;letter-spacing:0px;word-spacing:0px;writing-mode:lr-tb;text-anchor:start;display:inline;fill:#000000;fill-opacity:1;stroke:none;stroke-width:0.26458332" | |||
| xml:space="preserve"><tspan | |||
| style="font-size:7.76111126px;stroke-width:0.26458332" | |||
| y="156.9364" | |||
| x="133.6673" | |||
| id="tspan7173-9-7" | |||
| sodipodi:role="line">c</tspan></text> | |||
| </g> | |||
| <g | |||
| transform="translate(-125.96169,45.558965)" | |||
| id="g8392-6"> | |||
| <circle | |||
| r="4.0497961" | |||
| cy="154.90707" | |||
| cx="135.72226" | |||
| id="circle823-9-3" | |||
| style="display:inline;opacity:1;fill:#ffffff;fill-opacity:1;stroke:#000000;stroke-width:0.35277778;stroke-miterlimit:10;stroke-dasharray:none;stroke-opacity:1" /> | |||
| <text | |||
| id="text7175-0-4-2" | |||
| y="156.9364" | |||
| x="133.6673" | |||
| style="font-style:normal;font-variant:normal;font-weight:normal;font-stretch:normal;font-size:7.76111126px;line-height:1.25;font-family:'Trebuchet MS';-inkscape-font-specification:'Trebuchet MS, Normal';font-variant-ligatures:normal;font-variant-caps:normal;font-variant-numeric:normal;font-feature-settings:normal;text-align:start;letter-spacing:0px;word-spacing:0px;writing-mode:lr-tb;text-anchor:start;display:inline;fill:#000000;fill-opacity:1;stroke:none;stroke-width:0.26458332" | |||
| xml:space="preserve"><tspan | |||
| style="font-size:7.76111126px;stroke-width:0.26458332" | |||
| y="156.9364" | |||
| x="133.6673" | |||
| id="tspan7173-9-7-0" | |||
| sodipodi:role="line">e</tspan></text> | |||
| </g> | |||
| <path | |||
| style="fill:none;stroke:#000000;stroke-width:0.26458332px;stroke-linecap:butt;stroke-linejoin:miter;stroke-opacity:1" | |||
| d="m -20.78869,160.92857 22.1116066,0.75595" | |||
| id="path4607" | |||
| inkscape:connector-curvature="0" /> | |||
| <path | |||
| style="fill:none;stroke:#000000;stroke-width:0.26458332px;stroke-linecap:butt;stroke-linejoin:miter;stroke-opacity:1" | |||
| d="M 5.4806548,166.97619 2.6458333,177.9375" | |||
| id="path4609" | |||
| inkscape:connector-curvature="0" /> | |||
| <path | |||
| style="fill:none;stroke:#000000;stroke-width:0.26458332px;stroke-linecap:butt;stroke-linejoin:miter;stroke-opacity:1" | |||
| d="m -12.095238,187.3869 7.1815475,-2.83482" | |||
| id="path4611" | |||
| inkscape:connector-curvature="0" /> | |||
| <path | |||
| style="fill:none;stroke:#000000;stroke-width:0.26458332px;stroke-linecap:butt;stroke-linejoin:miter;stroke-opacity:1" | |||
| d="m 3.7797619,187.95387 3.2127975,7.18154" | |||
| id="path4613" | |||
| inkscape:connector-curvature="0" | |||
| sodipodi:nodetypes="cc" /> | |||
| <path | |||
| style="fill:none;stroke:#000000;stroke-width:0.26458332px;stroke-linecap:butt;stroke-linejoin:miter;stroke-opacity:1" | |||
| d="m -24.568452,165.84226 5.291667,17.57589" | |||
| id="path4615" | |||
| inkscape:connector-curvature="0" | |||
| sodipodi:nodetypes="cc" /> | |||
| <path | |||
| style="fill:none;stroke:#000000;stroke-width:0.26458332px;stroke-linecap:butt;stroke-linejoin:miter;stroke-opacity:1" | |||
| d="m -12.284226,191.35565 16.8199402,7.74851" | |||
| id="path4636" | |||
| inkscape:connector-curvature="0" /> | |||
| </g> | |||
| </svg> | |||
+ 4235
- 0
courses/lab06_data/OMIM.csv
文件差异内容过多而无法显示
查看文件
+ 374
- 0
courses/lab06_graphes.Rmd
查看文件
| @@ -0,0 +1,374 @@ | |||
| --- | |||
| title: "Graphes" | |||
| author: "Maxime Wack" | |||
| date: "19/11/2019" | |||
| output: | |||
| xaringan::moon_reader: | |||
| css: ['default','css/my_style.css'] | |||
| lib_dir: libs | |||
| seal: false | |||
| nature: | |||
| ratio: '4:3' | |||
| countIncrementalSlides: false | |||
| self-contained: true | |||
| beforeInit: "addons/macros.js" | |||
| highlightLines: true | |||
| pdf_document: | |||
| seal: false | |||
| --- | |||
| ```{r setup, include=FALSE} | |||
| library(tidyverse) | |||
| library(DT) | |||
| library(knitr) | |||
| opts_chunk$set(echo = TRUE, | |||
| ## fig.asp= .5, | |||
| message = F, | |||
| warning = F) | |||
| options(DT.options = list(paging = F, | |||
| search = F, | |||
| info = F)) | |||
| datatable <- partial(datatable, rownames = F) | |||
| ``` | |||
| class: center, middle, title | |||
| # UE Visualisation | |||
| ### 2019-2020 | |||
| ## Dr. Maxime Wack | |||
| ### AHU Informatique médicale | |||
| #### Hôpital Européen Georges Pompidou, </br> Université de Paris | |||
| --- | |||
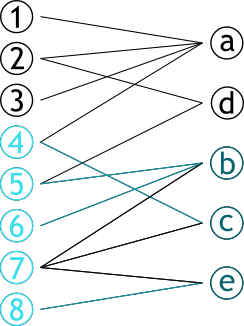
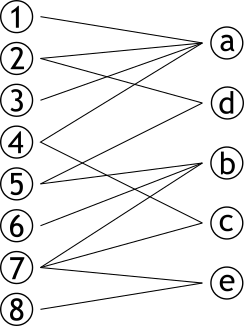
| # Graphe bipartite | |||
| .pull-left[ | |||
| ### Graphe contenant deux sets de sommets **complèment déconnectés** | |||
| ### Dit aussi 2-coloriable | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
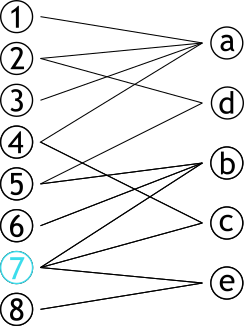
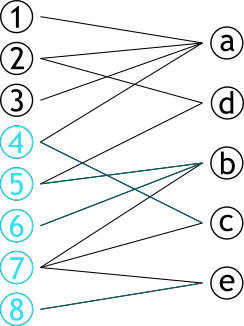
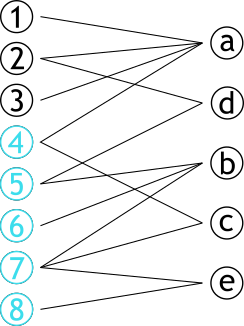
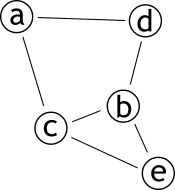
| # Projection | |||
| .pull-left[ | |||
| ### Méthode permettant de créer **deux** graphes | |||
| ### Les sommets d'une partie sont connectés s'ils partagent un sommet d'une autre partie | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
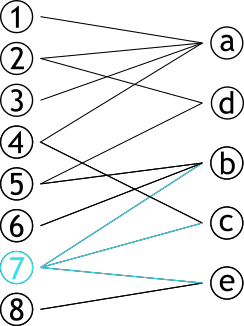
| # Projection | |||
| .pull-left[ | |||
| ### Méthode permettant de créer **deux** graphes | |||
| ### Les sommets d'une partie sont connectés s'ils partagent un sommet d'une autre partie | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
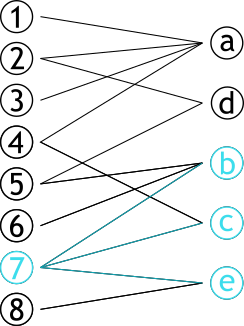
| # Projection | |||
| .pull-left[ | |||
| ### Méthode permettant de créer **deux** graphes | |||
| ### Les sommets d'une partie sont connectés s'ils partagent un sommet d'une autre partie | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
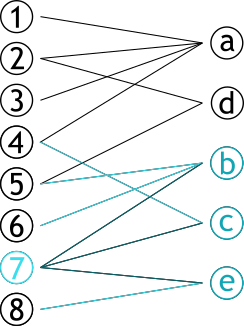
| # Projection | |||
| .pull-left[ | |||
| ### Méthode permettant de créer **deux** graphes | |||
| ### Les sommets d'une partie sont connectés s'ils partagent un sommet d'une autre partie | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
| # Projection | |||
| .pull-left[ | |||
| ### Méthode permettant de créer **deux** graphes | |||
| ### Les sommets d'une partie sont connectés s'ils partagent un sommet d'une autre partie | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
| # Projection | |||
| .pull-left[ | |||
| ### Méthode permettant de créer **deux** graphes | |||
| ### Les sommets d'une partie sont connectés s'ils partagent un sommet d'une autre partie | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
| # Projection | |||
| .pull-left[ | |||
| ### Méthode permettant de créer **deux** graphes | |||
| ### Les sommets d'une partie sont connectés s'ils partagent un sommet d'une autre partie | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
| class:center | |||
| # Projection | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
| class:center | |||
| # Projection | |||
| .pull-left[ | |||
|  | |||
| ] | |||
| .pull-right[ | |||
|  | |||
| ] | |||
| --- | |||
| class:center | |||
| # Projection | |||
| .pull-c1[ | |||
|  | |||
| ] | |||
| .pull-c2[ | |||
|  | |||
| ] | |||
| -- | |||
| .pull-c3[ | |||
|  | |||
| ] | |||
| --- | |||
| # OMIM | |||
| *Online Mendelian Inheritance in Men* | |||
| Base de données d'associations connues gène ↔ phénotype | |||
| https://omim.org | |||
| https://maximewack.com/files/OMIM.csv | |||
| --- | |||
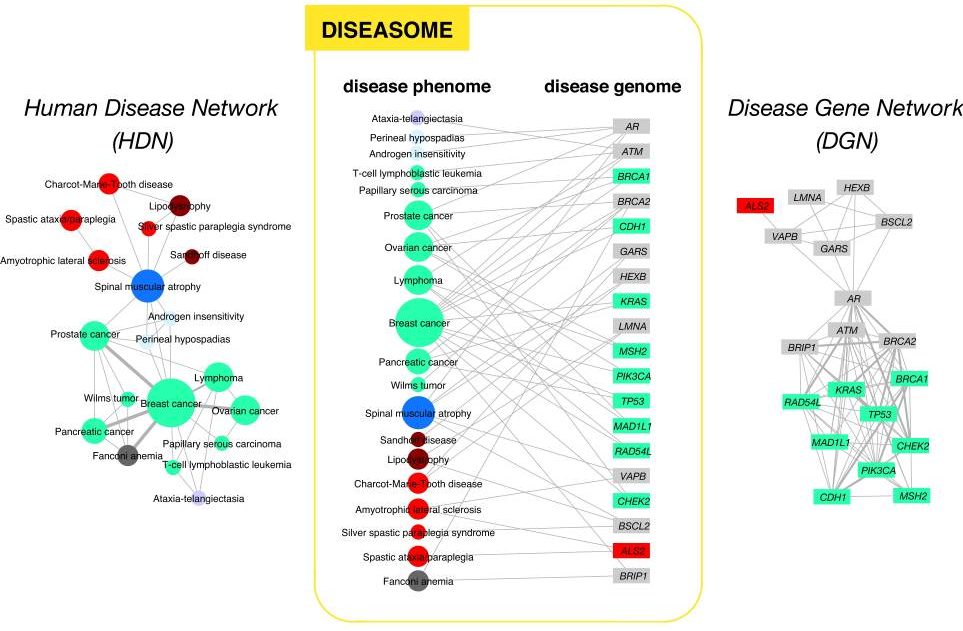
| class: center | |||
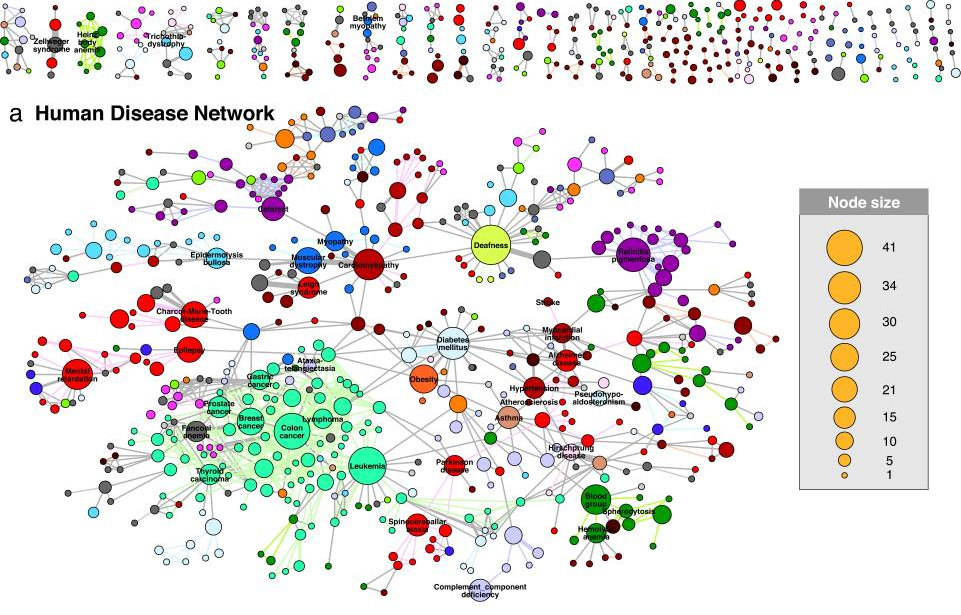
| # The Human Disease Network | |||
| https://www.ncbi.nlm.nih.gov/pubmed/17502601 | |||
|  | |||
| --- | |||
| class: center | |||
| # The Human Disease Network | |||
| https://www.ncbi.nlm.nih.gov/pubmed/17502601 | |||
|  | |||
| --- | |||
| # Librairies | |||
| ```{r libs} | |||
| library(igraph) | |||
| library(ggraph) | |||
| library(tidyverse) | |||
| ``` | |||
| --- | |||
| class:center,middle | |||
| # igraph | |||
| --- | |||
| # Créer des graphes | |||
| ```{r create graph, eval = F} | |||
| # Graphe sans arête | |||
| graph.empty(n = 10, directed = T) | |||
| # Graphe complètement connecté | |||
| graph.full(n = 10, directed = F, loops = F) | |||
| # Graphe en étoile | |||
| graph.star(n = 10, mode = "out") | |||
| ``` | |||
| --- | |||
| # Chargement du graphe | |||
| ```{r load} | |||
| read_csv("lab06_data/OMIM.csv") -> OMIM | |||
| ``` | |||
| ```{r load show, echo = F} | |||
| OMIM %>% | |||
| slice(70:80) %>% | |||
| datatable | |||
| ``` | |||
| --- | |||
| # Chargement du graphe | |||
| ```{r graph data frame} | |||
| graph.data.frame(OMIM, directed = F) -> graphe | |||
| ``` | |||
| ```{r graph data frame show, echo = F} | |||
| graphe | |||
| ``` | |||
| --- | |||
| # Informations sur le graphe | |||
| ### Sommets | |||
| ```{r vcount} | |||
| vcount(graphe) | |||
| ``` | |||
| ### Arêtes | |||
| ```{r ecount} | |||
| ecount(graphe) | |||
| ``` | |||
| --- | |||
| # Informations sur le graphe | |||
| ### Dirigé ? | |||
| ```{r directed} | |||
| is.directed(graphe) | |||
| ``` | |||
| ### Voisins d'un sommet | |||
| ```{r neighbors} | |||
| neighbors(graphe, V(graphe)[2019]) | |||
| ``` | |||
| --- | |||
| # Projection | |||
| ```{r projection} | |||
| # Établir les types (gène, phénotype) | |||
| V(graphe)$type <- bipartite.mapping(graphe)$type | |||
| # Créer les projections | |||
| projs <- bipartite.projection(graphe) | |||
| # Séparer les projections en deux graphes | |||
| HDN <- projs$proj1 | |||
| HGN <- projs$proj2 | |||
| ``` | |||
| --- | |||
| # HDN | |||
| ```{r HDN} | |||
| HDN | |||
| ``` | |||
| --- | |||
| # HGN | |||
| ```{r HGN} | |||
| HGN | |||
| ``` | |||
| --- | |||
| # Décomposition en sous-graphes | |||
| ```{r decompos} | |||
| HDN %>% | |||
| decompose -> diseases | |||
| ``` | |||
| --- | |||
| # ggraph | |||
| ```{r visu} | |||
| graph_one <- function(graph) | |||
| { | |||
| ggraph(graph) + | |||
| geom_edge_diagonal() + | |||
| geom_node_label(aes(label = name)) | |||
| } | |||
| ``` | |||
| --- | |||
| # Filtrer sous-graphes < 10 sommets | |||
| ```{r sous-graphes par sommets} | |||
| diseases %>% | |||
| keep(map_dbl(diseases, vcount) >= 10) %>% | |||
| map(graph_one) -> plots | |||
| ``` | |||
| --- | |||
| # Malformations cardiaques | |||
| ```{r cardiaques} | |||
| plots[[5]] | |||
| ``` | |||
| --- | |||
| # Surdités | |||
| ```{r surdités} | |||
| plots[[9]] | |||
| ``` | |||
| --- | |||
| # Ostéogénèses imparfaites | |||
| ```{r Ostéogénèses} | |||
| plots[[12]] | |||
| ``` | |||
| --- | |||
| # Export | |||
| ```{r export, eval = F} | |||
| write.graph(HDN, file = "diseases.graphml", format = "graphml") | |||
| write.graph(HGN, file = "genes.graphml", format = "graphml") | |||
| ``` | |||